This tutorial shows you how to extend LabKey Biologics to include purification workflows. To help you answer questions such as:
- Which purification system (i.e., some combination of purification elements) is more efficient?
- How does heat stress effect the quality of the drug substance?
Assume the following general workflow steps:
- You have batches of broth from various expression systems. Purification of these samples produces a usable drug substance
- A variety of purification regimens are available: different column designs, resins, AEX processes, and other experimental factors.
- The drug substance is subjected to a variety of stress tests, such as heat tests, oxidation tests, etc.
- Finally, MS2 experiments are performed, which rate the experimental factors (the purification regimens and stress tests) based on observed peptide modifications.
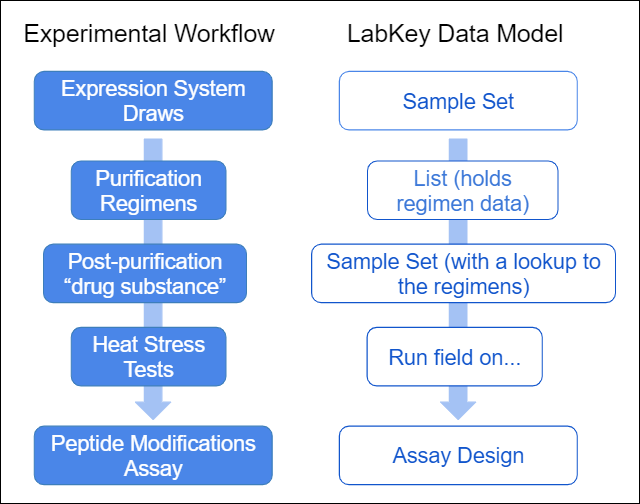
The following data model provides the LabKey data structures to capture the phases of experimental workflow.
The following tutorial gives you step by step instructions for capturing this experimental workflow in LabKey Biologics.
Download
- Download the 4 example files below, used for assembling the workflow.
Set Up Folder and Dashboard
- On LabKey Server, navigate to a project where you have admin control.
- Navigate to a subfolder or make a subfolder called "PurificationWorkflow". Below we assume a blank folder of type Assay, but you work in any folder type, for example a Biologics folder.
- Enter > Page Admin Mode.
- On the left side, add the Web Parts:
- Assay List
- Sample Types
- Lists
- Click Exit Admin Mode.
List
- Create a List named 'PurificationRegimens'.
- Infer fields, and import data from, the following data file: ListPurificationRegimens.txt
- Select RegimenName as the primary key.
Sample Types
- Create a Sample Type named 'PrePurificationSamples'.
- Add the following fields:
- Volume (Decimal)
- Volume_units (Text)
- Import the following data file: SamplesPrePurification.txt
- Create a Sample Type named 'PostPurificationSamples'.
- Add the following fields:
- Volume (Decimal)
- Volume_units (Text)
- PurificationRegime (Lookup) -- point the lookup column to the List PurificationRegimens.
- Import the following data file: SamplesPostPurification.txt
Assay Design
- Create a new Assay Design called "PeptideModifications".
- Remove the 6 default fields, in particular:
- Remove the 2 Batch Fields.
- Remove the 4 Result Fields.
- Infer the Results Fields from the following file: AssayPeptideModifications1.txt. This should give you the following 6 fields:
- Protein (Text)
- Location (Text)
- Modification (Text)
- Experiment (Text)
- Intensity (Decimal)
- Rel. Intensity (Decimal)
- Add the following Run Fields:
- SampleId (Sample) - for the Sample Options, set Sample lookup to PostPurificationSamples
- StressTemperature (Decimal)
- Save the design.
- Go the Runs or Result grids (which are currently empty) and click Import Data.
- Set the Run fields as follows:
- SampleId - Select some target sample from the dropdown, especially one of the PostPurificaiton samples.
- StressTemperature - Enter a decimal value for the stress temperature, for example, 5, 25, or 40.
- Import data from the same data file used above: AssayPeptideModifications1.txt
- Import additional assay data in these files, selecting different SampleIds and Heat tests as you import:
Grid Modification
- Go to the Assay result grid.
- Open Grid views > Customize Grid
- Open the nodes: Run > SampleId > PurificationRegimen.
- Select the fields: RegimenName, AEXConductivity, ColProtein, Column, Resin.
- Save this grid as the Default and select "Make this grid view available to all users".
- Now you have a grid which you can query upon to compare the performance of the different regimens and stress tests.
 The following data model provides the LabKey data structures to capture the phases of experimental workflow.
The following data model provides the LabKey data structures to capture the phases of experimental workflow. The following tutorial gives you step by step instructions for capturing this experimental workflow in LabKey Biologics.
The following tutorial gives you step by step instructions for capturing this experimental workflow in LabKey Biologics.