| Jon (LabKey DevOps) responded: |
2018-01-16 10:58 |
Hi Tomas,
Do you recall how long ago you upgraded to 17.2 Community r54372? Are you still on that version right now or have you upgraded to 17.3 since then?
Was the slowness immediate after the upgrade or was it only recently that you noticed the slowness?
Is it possible for you to send us your logs and indicate where in the logs it demonstrates this behavior? This will help with the investigation.
Regards,
Jon |
| |
| tvaisar responded: |
2018-01-16 11:10 |
Jon,
reviewing the upload logs much closer, it seems that the slowdown did not start abruptly after the upgrade, but was slowly creeping up even before. However, after the upgrade it seemed to jump up more.
I have upgraded to 17.3 last week in hope of improvement but it stayed the same.
What can I look at to try to troubleshoot?
In the last upgrade of the PostgrSQL database I noticed that it is at 13 GB. Could that have something to do with it?
Tomas |
| |
| Jon (LabKey DevOps) responded: |
2018-01-16 22:18 |
Hi Tomas,
The best place to start troubleshooting is to use log files that generated from the import job to see the log output to identify where the slowness is coming from. This would narrow down the source of the bottleneck when processing the files.
Can you send us the logs so we can take a look at them?
Regarding your database, you can run a VACUUM ANALYZE on the database. Here is some info on the VACUUM and the ANALYZE commands to help you get started: https://wiki.postgresql.org/wiki/Introduction_to_VACUUM,_ANALYZE,_EXPLAIN,_and_COUNT
Regards,
Jon |
| |
| tvaisar responded: |
2018-01-16 22:43 |
Jon,
I could send you the logs, but perhaps what may be easier is if I gave you access to our Labkey server.
Let me know what you want me to do.
In the meantime, I am going to look at the VACUUM ANALYZE.
Tomas |
| |
| Jon (LabKey DevOps) responded: |
2018-01-17 14:36 |
Hi Tomas,
Please send us the logs at your earliest convenience.
Regards,
Jon |
| |
| tvaisar responded: |
2018-01-17 14:49 |
Jon,
attached are several interact-ProteinProphet upload log files. I too examples from a period of over a year.
Tomas |
|
| |
| Jon (LabKey DevOps) responded: |
2018-02-02 13:33 |
Checking the logs, there is an unusual gap of time seen in some of them.
The following files show no lag time in processing:
161118_interact-161108BRIT1D_H1_08.prot.xml.log
170602_interact-170523FACSDIO_L21G10.prot.xml.log
170927_interact-170924LevRosnerA4.prot.xml.log
These logs show a lag:
171120_interact-171114LevIFNGR_11.prot.xml.log - Lag time occurs at the point when it looks for a 'sp|' prefix:
20 Nov 2017 07:21:47,963 INFO : Importing MS/MS results is 100% complete
20 Nov 2017 07:21:48,517 INFO : Spectra were not imported: pep.xml file included "pipeline, import spectra = no" or "pipeline, load spectra = no" setting.
20 Nov 2017 07:21:48,529 DEBUG: 131.79 seconds (2.20 minutes) to import peptide search results
20 Nov 2017 07:21:48,530 INFO : Starting to update SeqId column
20 Nov 2017 07:23:46,320 INFO : Set SeqId values for 29666 peptides based on exact protein name match for FASTA id 17
20 Nov 2017 07:24:04,584 INFO : Set SeqId values for 40 peptides based on protein identifier match from SwissProt database for FASTA id 17
20 Nov 2017 07:24:04,704 INFO : Set SeqId values for 0 peptides based on trailing FASTA header line for FASTA id 17
20 Nov 2017 07:39:32,468 INFO : Set SeqId values for 0 peptides based on 'sp|' prefix name match for FASTA id 17
20 Nov 2017 07:39:32,469 DEBUG: 1063.94 seconds (17.73 minutes) to update SeqId column
20 Nov 2017 07:39:32,469 INFO : Starting to update SequencePosition column
171219_interact-171212BelaAExp4A1_Q.prot.xml.log - Shows the same point when it lags:
19 Dec 2017 05:07:50,348 INFO : Importing MS/MS results is 100% complete
19 Dec 2017 05:07:50,794 INFO : Spectra were not imported: pep.xml file included "pipeline, import spectra = no" or "pipeline, load spectra = no" setting.
19 Dec 2017 05:07:50,800 DEBUG: 203.12 seconds (3.39 minutes) to import peptide search results
19 Dec 2017 05:07:50,800 INFO : Starting to update SeqId column
19 Dec 2017 05:14:31,215 INFO : Set SeqId values for 10376 peptides based on exact protein name match for FASTA id 18
19 Dec 2017 05:15:06,420 INFO : Set SeqId values for 0 peptides based on protein identifier match from SwissProt database for FASTA id 18
19 Dec 2017 05:15:06,820 INFO : Set SeqId values for 0 peptides based on trailing FASTA header line for FASTA id 18
19 Dec 2017 05:56:02,385 INFO : Set SeqId values for 0 peptides based on 'sp|' prefix name match for FASTA id 18
19 Dec 2017 05:56:02,385 DEBUG: 2891.58 seconds (48.19 minutes) to update SeqId column
19 Dec 2017 05:56:02,385 INFO : Starting to update SequencePosition column
180112_interact-180108LevLysE1.prot.xml.log - Same thing there as well:
12 Jan 2018 23:30:21,952 INFO : Importing MS/MS results is 100% complete
12 Jan 2018 23:30:22,026 INFO : Spectra were not imported: pep.xml file included "pipeline, import spectra = no" or "pipeline, load spectra = no" setting.
12 Jan 2018 23:30:22,032 DEBUG: 32.70 seconds to import peptide search results
12 Jan 2018 23:30:22,032 INFO : Starting to update SeqId column
12 Jan 2018 23:31:40,062 INFO : Set SeqId values for 33027 peptides based on exact protein name match for FASTA id 18
12 Jan 2018 23:32:00,869 INFO : Set SeqId values for 49 peptides based on protein identifier match from SwissProt database for FASTA id 18
12 Jan 2018 23:32:00,973 INFO : Set SeqId values for 0 peptides based on trailing FASTA header line for FASTA id 18
12 Jan 2018 23:48:16,908 INFO : Set SeqId values for 0 peptides based on 'sp|' prefix name match for FASTA id 18
12 Jan 2018 23:48:16,908 DEBUG: 1074.88 seconds (17.91 minutes) to update SeqId column
12 Jan 2018 23:48:16,908 INFO : Starting to update SequencePosition column |
| |
| tvaisar responded: |
2018-02-02 13:49 |
Hi Jony,
Yes, that is what I noticed. However, I am not sure why would that be.
What you are saying is, it is at the point where it is looking through the FASTA file for the source database prefix? Or when it is looking through the pep.xml file?
Is there a way I can troubleshoot this?
Tomas |
| |
| Jon (LabKey DevOps) responded: |
2018-02-03 01:00 |
Hi Tomas,
Based on the log info, it appears that the import is having issue due to not being able to match some of the protein names in the pepXML file to the FASTA file.
You should be able to identify those that don't match by going to the peptide list, customizing the view, then add the Search Engine Protein > Best Name field. Then you would filter that column for blank values.
Give that a try and let us know how that goes.
Regards,
Jon |
| |
| tvaisar responded: |
2018-02-03 09:16 |
Jon,
I tried it with the 171219_interact-171212BelaAExp4A1_Q.prot.xml and I am not finding any blank values.
Just to make sure I am doing what you suggest - I go to the MS2 Runs page, choose this prot.xml file to get to the page where I have Run Overview, View, and Proteins and Peptides. In Proteins and Peptides section I go to Customize Grid and in available fields I choose Search Engine Protein > Best Name, go to View Grid (this results in pivoted view showing Best Name and collapsed peptides view). I filter that view to "Is Blank". And there is nothing.
There is also predefined view "SearchEngineProtein" in which I filtered Best Name to "Is Blank" and again - no blanks show up.
Next thing to try?
Tomas |
| |
| Jon (LabKey DevOps) responded: |
2018-02-13 13:34 |
Hi Tomas,
Can you try looking for protein names that aren't linked?
Take a look at the highlighted rows under the Protein column in the screenshot.
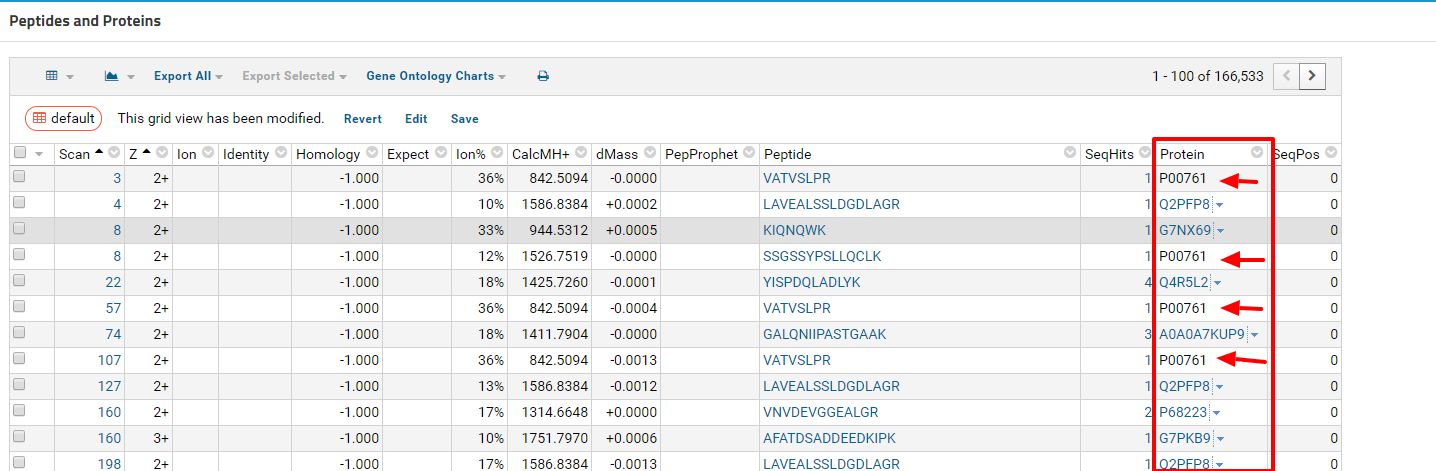
Regards,
Jon |
|
| |
| tvaisar responded: |
2018-02-13 14:50 |
OK,
I think we are getting somewhere - I do DECOY searches and there are some decoy peptides which are showing up and they do not have the link.
Could that be it?
Screenshot attached.
How do you deal with Decoy search results - I use the utility in Comet which generates the DECOY database on the fly so it is not part of the FASTA database file.
Tomas |
|
| |
| Jon (LabKey DevOps) responded: |
2018-02-16 23:07 |
Hi Tomas,
Those decoy proteins would explain the slow processing since it's trying to make a match to the FASTA file and fails.
When it comes to Comet searches, we don't really have a better way to do this currently or have any feasible workarounds.
I've setup bug https://www.labkey.org/home/Developer/issues/issues-details.view?issueId=33269 on this for further investigation, but this may take some time to fix and get released.
Regards,
Jon |
| |
| Jon (LabKey DevOps) responded: |
2024-02-29 10:38 |
|
| |