Table of Contents |
guest 2025-06-14 |
LabKey Biologics - Videos
General Set Up and Terminology
Summary
Terminology
Building Biologics
Entity Registration
Search the Registry
Registry Dashboard
Register a New Nucleotide Sequence
Register a New Protein Sequence
Protein Sequence Annotations
Register a New Molecule
Molecule Sets and Molecular Species
Vectors, Constructs, and Cell LinesExpression Systems
Use the Registry API
Configuring Grids, Detail Views, and Entry Forms
Sample Management
Samples and Sample Sets Within LabKey Server
Sample Sets and Detail Pages
Generate Samples
Assay Management
Set Up Assays
Media Registration
Registering Ingredients and Raw Materials
Registering Mixtures (Recipes)
Registering Batches
Workflow Management
Workflow
Service Request Tracker Set Up
LabKey Biologics
- Easily and uniquely register all biological entities individually or in bulk.
- Track and query the lineage of samples throughout many generations of derivation.
- Integrate biological entity information with sample information and downstream assay data used to evaluate therapeutic properties.
- Manage and monitor the execution of laboratory tasks.
Video Resources
- LabKey Biologics Overview
- Intro to LabKey Biologics
- Quick Look: Navigating Entities and Connections Between Entity Types
- Quick Look: Registering Nucleotide and Protein Sequences with LabKey Biologics
- Quick Look: Assays in LabKey Biologics
- Quick Look: Registering Media in LabKey Biologics
- Quick Look: Registering Molecules in LabKey Biologics
- Quick Look: Managing Assay Requests in LabKey Biologics
- Quick Look: Customizing Sample ID Structures and Generating Sample IDs in LabKey Biologics
- Quick Look: Automatic Generation of Molecular Species
General Set Up and Terminology
Entity Registration
- Search the Registry
- Registry Dashboard
- Register a New Nucleotide Sequence
- Register a New Protein Sequence
- Register a New Molecule
- Molecular Species and Molecule Sets
- Vectors, Cell Lines, and Constructs
- Registry API
- Configuring Columns Seen in Grids and Detail Views
Sample Management
Assay Management
Media Registration
- Registering Ingredients and Raw Materials
- Registering Mixtures (Recipes)
- Registering Mixture Batches
Workflow Management
- Workflow
- Assay Request Workflow - Administrator Documentation
- Service Request Tracker Set Up - Administrator Documentation
Related Topics
LabKey Biologics - Videos
The following Biologics videos are available on the LabKey YouTube channel:
- LabKey Biologics Overview
- Intro to LabKey Biologics
- Quick Look: Navigating Entities and Connections Between Entity Types
- Quick Look: Registering Nucleotide and Protein Sequences with LabKey Biologics
- Quick Look: Assays in LabKey Biologics
- Quick Look: Registering Media in LabKey Biologics
- Quick Look: Registering Molecules in LabKey Biologics
- Quick Look: Managing Assay Requests in LabKey Biologics
- Quick Look: Customizing Sample ID Structures and Generating Sample IDs in LabKey Biologics
- Quick Look: Automatic Generation of Molecular Species
General Set Up and Terminology
- Summary - An overview of the user interface.
- Terminology - Terms used throughout LabKey Biologics.
- Building Biologics - Information for developers who are building LabKey Biologics from source code.
Summary
LabKey Biologics: Home Page
The Home page provides a search box and different portals into different aspects of the data:
- Search - Search for entities in the registry.
- Registry - Browse all of the entities in the registry.
- Assays - Assay results for candidate molecules.
- Samples - A dashboard for tracking samples.
- Media - A dashboard for ingredients, media, mixtures, and recipes.
- Workflow - Request assay runs on samples, creation of media batches, and track progress.

Dashboard Navigation
When viewing a dashboard, navigate around the registry using the menu bar at the top of the page.

Hover over a menu item for sub-menus.

Registry
The Registry dashboard shows all of the unique entities that have been added, such as Molecules, Expression Systems, Cell Lines, etc. Click a tile to learn more or register new entities:

Learn more about the Registry section and the relationships between entities in Registry Dashboard.
Samples
The Samples dashboard shows a grid of samples recently added to the registry. Rows represent sample sets and columns, months. Darker panels indicate more samples for that month.

To learn more about the Samples features, see Sample Sets and Detail Pages.
Assays
The Assays dashboard shows data broken down by type and date. Similar to the Samples dashboard, darker tiles in the grid indicate a greater quantity of data for that assay in that month.

Media
From the Media dashboard, you add and manage ingredients, raw materials, mixtures, and batches.
See Registering Ingredients and Raw Materials for more information.
Workflow
The Workflow dashboard contains:
- Various reports on assay requests
- A grid of assay requests currently in the pipeline

More information can be found in the topic: Workflow
Related Topics
Learn more about each aspect of the registry in these detailed topics:
Terminology
Diagram of Entity Relationships

Molecule
Composed of a mixture of protein sequences, nucleotide sequences, chemistry elements, and other molecules. Generally, "molecule" refers to the target entity.
| Example Molecules |
|---|
| Molecule W = 1 (protein sequence A) |
| Molecule X = 1 (protein sequence B) + 2 (protein sequence C) |
| Molecule Y = 1 (nucleotide sequence D) |
| Molecule Z = 2 (nucleotide sequence E) + 2 (protein sequence F) + 2 (molecule W) |
Molecule Set
The set of molecules that only differ from one another in terms of their leader sequences. See Vectors, Constructs, and Cell LinesExpression Systems.
Molecular Species
After protein expression and other processes, all the entities that are detected for a particular molecule (different cleavage sites, post-translational modifications, genomic drift). See Vectors, Constructs, and Cell LinesExpression Systems.
Protein Sequence
Single sequence comprised of amino acids (20 different amino acids).
| Example Protein Sequences |
|---|
| protein sequence A = ACELKYHIKL CEEFGH |
| protein sequence B = HIKLMSTVWY EFGHILMNP |
Nucleotide Sequence
A single sequence comprised of nucleic acids, can be either DNA (A, T, G, C) or RNA (A, U, G, C).
| Example Nucleotide Sequences |
|---|
| nucleotide sequence A (DNA) = AGCTGCGTGG GCACTCACGCT |
| nucleotide sequence B (RNA) = AGCUGUUGCA GCUUACAUCGU |
Along with being a component of a molecule, DNA sequences can be designed that encode for specific protein sequences when transferred into a cell line to create an expression system.
Vector
DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell. The most common type of vector is a plasmid - a long double-stranded section of DNA that has been joined at the ends to circularize it. In molecular biology, generally, these plasmids have stretches of DNA somewhere in their makeup that allow for antibiotic resistance of some sort, providing a mechanism for selecting for cells that have been successfully transfected with the vector.
Construct
A vector (generally a plasmid) that has had a stretch of DNA inserted into it. Generally, this DNA insertion encodes for a protein of interest that is to be expressed.
Cell Line
A host cell line is transfected with a Construct, bringing the new DNA into the machinery of the cell. These transfected cells (called an Expression System) are then processed to select for cells that have been successfully transfected and grown up, allowing for the production of cells that have the construct and are manufacturing the protein of interest. At times, these transfections are transient and all of the cells are used in the process. Other times, a new stable cell line (still a Cell Line) is produced that can continue to be used in the future.
Expression System
A combination of a Construct and a host cell
Related Topics
Building Biologics
General Development Set Up
Complete the general build instructions.
Add Biologics Modules
Get the biologics and related modules from github:
> cd server/optionalModules
> git clone https://github.com/LabKey/biologics.git
> git clone https://github.com/LabKey/assayRequest.git
> git clone https://github.com/LabKey/recipe.git
> git clone https://github.com/LabKey/inventory.git
Add nodejs and npm
To build the biologics module, you will need to install nodejs and npm.
https://nodejs.org/en/download/
Gradle Set Up
Add the modules to the file settings.gradle in the project root to the end of the file:
include ":server:optionalModules:assayRequest"
include ":server:optionalModules:biologics"
include ":server:optionalModules:inventory"
include ":server:optionalModules:recipe"
In IntelliJ, hit the Gradle refresh button to have these new modules added to the IntelliJ project.
Build LabKey Server with Biologics
Build the server and verify the new modules are compiled:
> ./gradlew deployApp
Related Topics
Entity Registration
Search the Registry
You can search the Biologics Registry in the following ways:
- Registry-Wide Search - Search across all entities in the registry.
- Grid Search Bar - Search and filter a particular grid of data.
Registry-Wide Search
To search the entire repository, enter your search terms on the home page (as shown below). Alternatively, click Search on the top navigation bar.

Results are displayed as tiles, as shown below.

Click Advanced to filter by Entity Type. The dropdown is populated by the entity types known to the system.

Results will be filtered to only the entity type selected.

Grid Search
A search bar appears above all grid views.

You can enter multiple criteria in the search bar, adding any number of filters, keywords, and sorts.

- filter columns - Enter an expression consisting of a column, a comparator, and a value. The grid will display only those rows where the expression is true (for the selected column). The screenshot below shows the Structure Format column filtered for the value 'Fv'.

- keyword search - Keyword searches behave like substring searches across all of the rows in the table, not just the rows currently displayed in the browser. But the search is limited to only those columns that are currently visible in the browser. Hidden columns are not searched. To control which columns are visible and which are hidden, see Configuring Grids, Detail Views, and Entry Forms.

- sort columns - Sorts control the order in which rows are displayed. Below we have sorted the Name column from low to high values.

You can also control the sort order by clicking the triangle icons in the column headers.

Registry Dashboard
The Registry dashboard shows all of the unique entities that have been added, such as Molecules, Expression Systems, Cell Lines, etc.

Click Show entity relationships to see a graphical representation of how the entities are related to one another.

Click an entity tile to view data grids for that entity type. You can also directly select these entities from the Registry menu in the top bar.

Each grid shows the entities that have been registered. Click an entity name on a grid to see details. Some of these entity fields are eligible for editing, such as description or aliases, and any such edits are logged.
- Molecule Sets
- Molecules
- Molecular Species
- Protein Sequences
- Nucleotide Sequences
- Expression Systems
- Constructs
- Vectors
- Cell Lines
Register a New Nucleotide Sequence
This topic shows how to register a new nucleotide sequence using the graphical user interface. To register using the API, or to bulk import sequences from an Excel spreadsheet, see Use the Registry API.
To add a new nucleotide sequence to the registry:
From the header bar, select Registry > Nucleotide Sequences.

On the Nucleotide Sequences page, click Insert New.

Add details
- On the Register a new Nucleotide Sequence page, in the Details panel, enter the following:
- Description: an optional text description of the sequence.
- Alias: optional alternative names for the sequence.
- Nucleotide Sequence Parents: optional parent components. A related sequence the new sequence is derived from, for example, related as a mutation. You can select more than one parent.
- Sequence: Required field. The nucleotide sequence
- Annotations: Optional field. A comma separated list of annotation information:
- Name - a freeform name
- Category - region or feature
- Type - for example, Leader, Variable, Tag, etc.
- Start and End Positions are 1-based offsets within the sequence.
- Click Next.

Confirm
- To register the nucleotide and register the corresponding protein, click Finish and translate protein. This option will take you to the registry wizard for a new protein, prepopulating it with the protein sequence based on the nucleotide.
- To register the nucleotide and finish, click Finish.

Register a New Protein Sequence
This topic shows how to register a new protein sequence using the graphical user interface. To register using the API, or to bulk import sequences from an Excel spreadsheet, see Use the Registry API.
You can enter the Protein Sequence wizard in a number of ways:
- Via the nucleotide sequence wizard. When registering a nucleotide sequence, you have the option of continuing on to register the corresponding protein sequence.
- Via the header bar. Select Registry > Protein Sequences.
- Click Insert New (or use the "Insert" pulldown menu to select this option).

The wizard for registering a new protein sequence proceeds through four tabs:
Details
- Description: A text description of the sequence
- Alias: List one or more aliases. Type a name, enter when complete. Continue to add more as needed.
- Protein Sequence Parents: List optional parent component(s) for this sequence.
- Organism: Start typing the organism name or select from the pulldown menu.
Click Next to continue.
Sequence
On the sequence tab, you can translate a protein sequence from a nucleotide sequence as outlined below. If you prefer to manually enter a protein sequence from scratch click Manually add a sequence at the bottom.
- Nucleotide Sequence: Optional. The selection made here will populate the left-hand text box with the nucleotide sequence.
- Translation Frame: Required. The nucleotide sequence is translated into the protein sequence (which will be shown in the right-hand text box) by parsing it into groups of three. The selection of translation frame determines whether the first second or third nucleotide in the series 'heads' the first group of three. Options: 1,2,3.
- Sequence Length: This value is based on the selected nucleotide sequence.
- Nucleotide Start: This value is based on the nucleotide sequence and the translation frame.
- Nucleotide End: This value is based on the nucleotide sequence and the translation frame.
- Translated Sequence Length: This value is based on the nucleotide sequence and the translation frame.
- Protein Start: Specific the start location of the protein to be added to the registry.
- Protein End: Specific the end location of the protein to be added to the registry.
Click Next to continue.
Annotations
The annotations tab displays any matching annotations found in the annotation library. You can also add annotations manually at this point in the registration wizard. Editing is not allowed at this point, but you can edit annotations after the registration wizard is complete.
Suggested annotations can be “removed” by clicking the red icons in the grid panel. They can also be added back if the user changes their mind.

For complete details on using the annotation panel see Protein Sequence Annotations.
Properties
- Sequence
- Annotations: Optional field. The system will search for existing annotations for the protein. If none are found, you can enter an annotation manually using a comma separated list of items in the following order:
- Name: a freeform name
- Category: region or feature
- Type: for example, Leader, Variable, Tag, etc.
- Start and End Positions: 1-based offsets within the sequence
- Chain Format: select a chain format from the dropdown (start typing to filter the list of options). An administrator defines the set of options on the ChainFormats list. LabKey Biologics will attempt to classify the protein's chain format if possible.
- ε: the extinction coefficient
- Avg. Mass The average mass
- Num. S-S The number of disulfide bonds
- pI The isoelectric point
- Num Cys. The number of cysteine elements
Default or best guess values may prepopulate the wizard, but can be edited as needed.
Click Next to continue.
Confirm
The Confirm panel provides a summary of the protein about to be added to the registry.
Click Finish to add the protein to the registry.
Editing Protein Sequence Fields
Once you have defined a protein sequence, you can locate it on the lists and reopen to see the details. Some fields are eligible for editing. Those that are "in use" by the system or other entities cannot be changed. All edits are logged.
Protein Sequence Annotations
Annotation Detail Page
To view protein sequence annotations, go to Registry > Protein Sequences, and click a protein id in the Name column.
- The upper panel represents the sequence graphically, with different annotations called out with colored bars.
- The panel below provides annotation details in a grid view.
- The bottom panel provides an editor with two tabs for adding and editing annotations.

Interactions
Clicking an annotation’s color bar highlights its description in the grid below, For example, if you click the green bar for sequence position 36-49, that annotation’s details are highlighted in the grid below, and vice versa.

The annotations details grid can be sorted by any of the available columns. The grid will always default to Start in ascending order

The annotation editor has two tabs: Edit Annotation and Add Annotation. Adding or editing an annotation will refresh the grid and viewer to include your changes.
- Note that each field on the Add Annotation tab is required. The button at the bottom of the form will remain grayed out until all fields have been filled out.
- The Type dropdown is pre-populated with the items in the List "AnnotationTypes".

In order to edit an annotation, first select one from the grid or viewer. Once selected, click the Edit button at the bottom of the Edit Annotation tab.

This will enable the field inputs and action buttons:
- Cancel: Cancels the edit.
- Remove Annotation: Removes the annotation and deletes the details row from the grid.
- Save: Save any changes to the selected annotation. The Save button will become enabled only when changes have been made to the form.

The controls to the right of the Edit and Add tabs can be used to hide or show the position index and annotation color bars.
The sequence start and end points update to the currently selected annotation. The start and end points can be manually changed to any index values in the sequence, or can be reset to the full sequence by pressing the refresh button.
Clicking Copy sequence will copy the indicated segment to your clipboard for pasting (useful when registering a new protein sequence into the Registry).

A somewhat simplified version of the annotations viewer is also used within the protein sequence registration wizard.
Related Topics
Register a New Molecule
- Video: Registering a Molecule
- Related Topic: Register a New Nucleotide Sequence
- Related Topic: Register a New Protein Sequence
- Related Topic: Vectors, Constructs, and Cell LinesExpression Systems
This topic shows how to register a new molecule using the graphical user interface. To register using the API, or to bulk import molecules from an Excel spreadsheet, see Use the Registry API.
To add a new molecule to the registry:
- From the header bar, select Registry > Molecules.

- On the Molecules page, click Insert New.

Add details
- On the Register a new Molecule page, in the Add details panel, enter the following:
- Description: an optional text description of the molecule.
- Alias: optional alternative names for the molecule.
- Molecule Parents: optional parent molecules for the new molecule.

Click Next to continue.
Select Components
- On the Select components panel, search and select existing components of the new molecule.
- Using the appropriate radio button, search for the component of interest. You will see a partial details panel prior to adding the component.

- Once you have added a component, it will be shown as a panel the appropriate icon. Click to expand (the colors will reverse and chevron on the right will point down to indicate an expanded panel).
- You can copy the components sequence, or partial sequence, by clicking Copy Sequence. The sequence will be copied to your operating systems clipboard.
Click Next to continue.
Stoichiometry
LabKey Biologics will attempt to classify the structure format of the molecule's protein components, if possible. The structure format is based on the component protein chain formats.
On the Stoichiometry pane enter:
- Stoichiometry from each component
- S-S bonds
- Structure Format: Select a format from the pulldown list. The list is populated from the StructureFormat table.

Click Next to continue.
Confirm
On the Confirm pane, click Finish to add the molecule to the registry.

The new molecule will be added to the grid.

Related Topics
Molecule Sets and Molecular Species
Molecule Sets serve to group together Molecules (ex: antibodies or proteins) around common portions of protein sequences, once signal or leader peptides have been cleaved. Molecule Sets server as the "common name" for a set of molecules. Many Molecules may be grouped together in a single Molecule Set.
Molecular Species serve as alternate forms a given molecule. For example, a given antibody may give rise to multiple Molecular Species: one Species corresponding to the leaderless, or "mature", portion of the original antibody and another Molecular Species corresponding to its "mature, desK" (cleaved of signal peptides and heavy chain terminal lysine) form.
Both Molecular Species and Sets are calculated and created by the registry itself. Their creation is triggered when the user adds a Molecule to the registry. Users CAN also manually register Molecular Species, but generally do NOT register their own Molecule Sets. Detailed triggering and creation rules are described below.
When a molecule is added to the registry, the following additional entities are calculated and added, depending on the nature of the molecule. These additional entities can include:
- Molecular Species
- Molecule Sets
- Other Protein Sequences
Molecule Meriology
A molecule can be created (= registered) based on one or more of the following:
- a protein sequence
- a nucleotide sequence
- other molecules
Rules for Entity Calculation/Creation
When the molecule contains only protein sequences, then:
- A mature molecular species is created, consisting of the leaderless segments of the protein sequences, provided a leader portion is identifiable. The leader segment has to be:
- Have annotation start with residue #1
- The annotation Type is "Leader"
- The annotation Category is "Region"
- (If no leader portion is identifiable, then the species will be identical to the Molecule which has just been created.)
- Additionally, a mature desK molecular species is created (provided that there are terminal lysines on heavy chains).
- New protein sequences are created corresponding to any species created, either mature, mature des-K, or both. The uniqueness constraints imposed by the registry are in effect, so already registered proteins will be re-used, not duplicated.
- A molecule set is created, provided that the mature molecular species is new, i.e., is not the same (components and sequences) as any other mature molecular species of another molecule. If there is an already existing mature species in the registry, then the new molecule is associated with that set.
- Physical properties are not calculated.
- Molecular species are not created.
- A molecule set is created, which has only this molecule within it.

Aliases and Descriptions
When creating a Molecule, the auto-generated Molecule Set (if there is a new one) will have the same alias as the Molecule. If the new Molecule is tied to an already existing Molecule Set, the alias is appended with alias information from the new Molecule.
When creating a molecule, the auto-generated molecular species (both mature and mature desK) should have the same alias as the molecule. Similarly, when creating a molecular species from a molecule (manually), the alias field will pre-populate with the alias from the molecule.
For molecular species that are auto-generated, if it is creating new protein sequences (one or more) as the components of that molecular species, they have a Description:
- Mature of “PS-15”
- Mature, desK of “PS-16”
- Mature of “PS-17”
- Mature, desK of “PS-18”
Vectors, Constructs, and Cell LinesExpression Systems
Definitions
- Vectors are typically plasmids that can inject genetic material into cells. Must have specific nucleotide sequence.
- Constructs are Vectors which have been modified to include the genetic material intended for injection into the cell.
- Cell Lines are types of cells that can be grown in the lab.
- Expression Systems are cells line that has been injected with a construct.
Entry Forms
The biologics registry can capture Vectors, Constructs, Cell Lines, Expression Systems, and their relationships. To add these entities to the registry, use the entry forms at:
- Registry > Vectors > Insert New
- Registry > Constructs > Insert New
- Registry > Cell Lines > Insert New
- Registry > Expression Systems > Insert New
| Entity Type | Default Fields |
|---|---|
| Vector | Description Alias Sequence Selection Methods Vector Parents |
| Construct | Description Alias Vector Cloning Site Sequences Construct Paths |
| Cell Lines | Description Alias Expression System Stable Clonal Cell Line Parents |
| Expression Systems | Description Alias Host Cell Line Constructs Expression System Parents |
Use the Registry API
Register a Sequence or Molecule
Identity Service
When registering a sequence or molecule, use the "identity" of the sequence to determine uniqueness. External tools can use the following API to get the identity of a sequence prior to registration. To get the identity of a sequence use either the identity/get.api or identity/ensure.api. The ensure.api will create a new identity if the sequence hasn't been added yet.
To get or ensure the identity of a single nucleotide sequence:
LABKEY.Ajax.request({
url: LABKEY.ActionURL.buildURL("identity", "get.api"),
jsonData: {
items: [{
type: "nucleotide", data: "GATTACA"
}]
}
});To get or ensure the identity of a collection of sequences:
LABKEY.Ajax.request({
url: LABKEY.ActionURL.buildURL("identity", "get.api"),
jsonData: {
items: [{
type: "nucleotide", data: "GATTACA"
},{
type: "protein", data: "ELVISLIVES"
}]
}
});To get or ensure the identity of a molecule containing multiple protein sequences:
LABKEY.Ajax.request({
url: LABKEY.ActionURL.buildURL("identity", "get.api"),
jsonData: {
items: [{
type: "molecule", items: [{
type: "protein", data: "MAL", count: 2
},{
type: "protein", data: "SYE", count: 3
]}
}]
}
});Manual Entry
You can enter molecule, sequences, cell lines, etc, using registration wizards. For an example use of the wizard, see Register a New Nucleotide Sequence
Cut-and-Paste or Import from a File
The LabKey import data page can also be used to register new entities. For example, to register new nucleotide sequences:
- Go to the list of all entity types (the DataClasses web part).
- Click NucSequence.

- Click the Import Data button.
- Paste in a TSV file or upload an Excel file in the format:
| Description | protSeqId | translationFrame | translationStart | translationEnd | sequence |
|---|---|---|---|---|---|
| Anti_IGF-1 | PS-7 | 0 | 0 | 0 | caggtg... |
When importing a nucleotide sequence with a related protSeqId using the protein sequence's name, you will need to click the Import Lookups By Alternate Key checkbox on the Import Data page. The Name column may be provided, but will be auto-generated if it isn't. The Ident column will be auto-generated based upon the sequence.
To register new protein sequences:
- Go to the list of all entity types
- Click ProtSequence
- Click the Import Data button
- Paste in a TSV file or upload an Excel file in the format below.
| Name | Description | chainFormatId | organisms | sequence |
|---|---|---|---|---|
| PS-1 | PS1038-1 | 1 | ["human","mouse"] | MALWMRL... |
To register new molecules:
- Go to the list of all entity types
- Click Molecule
- Click the Import Data button
- Paste in data of the format:
| Description | structureFormatId | seqType | components |
|---|---|---|---|
| description | 1 | 1 | [{type: "nucleotide", name: "NS-1", stoichiometry: 3}] |
| description | 1 | 3 | [{type: "protein", name: "PS-1", stoichiometry: 2}, {type: "protein", ident: "ips:1234"}] |
| description | 1 | 3 | [{type: "chemical", name: "CH-1"}, {type: "molecule", name: "M-1"}] |
Note that the set of components is provided as a JSON array containing one or more sequences, chemistry linkers, or other molecules. The JSON object can refer to an existing entity by name (e.g "NS-1" or "PS-1") or by providing the identity of the previously registered entity (e.g., "ips:1234" or "m:7890"). If the entity isn't found in the database, an error will be thrown -- for now, all components must be registered prior to registering a molecule.
Register via Query API
The client APIs also can be used to register new entities.
Register Nucleotide Sequence
From your browser's dev tools console, enter the following to register new nucleotide sequences:
LABKEY.Query.insertRows({
schemaName: "exp.data",
queryName: "NucSequence",
rows: [{
description: "from the client api",
sequence: "gattaca"
},{
description: "another",
sequence: "cattaga"
}]
});Register Protein Sequence
To register new protein sequences:
LABKEY.Query.insertRows({
schemaName: "exp.data",
queryName: "ProtSequence",
rows: [{
name: "PS-100",
description: "from the client api",
chainFormatId: 1,
sequence: "ML"
}]
});Register Molecule
To register new molecules:
LABKEY.Query.insertRows({
schemaName: "exp.data",
queryName: "Molecule",
rows: [{
description: "from the client api",
structureFormatId: 1,
components: [{
type: "nucleotide", name: "NS-202"
}]
},{
description: "another",
structureFormatId: 1,
components: [{
type: "protein", name: "PS-1"
},{
type: "protein", name: "PS-100", count: 5
}]
}]
});Lineage, Derivation, and Samples
Parent/child relationships within an entity type are modeled using derivation. For example, the details page for this nucleotide sequence (NS-2) shows that another sequence (NS-34) has been derived from it.

To create new children, you can use the "experiment/derive.api" API, but it is still subject to change. The dataInputs is an array of parents each with an optional role. The targetDataClass is the LSID of the entity type of the derived datas. The dataOutputs is an array of children each with an optional role and a set of values.
LABKEY.Ajax.request({
url: LABKEY.ActionURL.buildURL("experiment", "derive.api"),
jsonData: {
dataInputs: [{
rowId: 1083
}],
targetDataClass: "urn:lsid:labkey.com:DataClass.Folder-5:NucSequence",
dataOutputs: [{
role: "derived",
values: {
description: "derived!",
sequence: "CAT"
}
}]
}
})Samples will be attached to an entity using derivation. Instead of a targetDataClass and dataOutputs, use a targetSampleSet and materialOutputs. For example:
LABKEY.Ajax.request({
url: LABKEY.ActionURL.buildURL("experiment", "derive.api"),
jsonData: {
dataInputs: [{
rowId: 1083
}],
targetSampleSet: "urn:lsid:labkey.com:SampleSet.Folder-5:Samples",
materialOutputs: [{
role: "sample",
values: {
name: "new sample!",
measurement: 42
}
}]
}
})Register Parents/Inputs
To indicate parents/inputs when registering an entity, use the columns "DataInputs/<DataClassName>" or "MaterialInputs/<SampleSetName>". The value in the column is a comma separated list of values in a single string. This works for both DataClass and SampleSet:
LABKEY.Query.insertRows({
schemaName: "exp.data",
queryName: "MyDataClass",
rows: [{
name: "blush",
"DataInputs/MyDataClass": "red wine, white wine"
}]
});Related Topics
Configuring Grids, Detail Views, and Entry Forms
Grids
- To control grid views, go the administration area by selecting Avatar > Admin.

- Find the grid view you wish to change and click it. For example, to change the grid view for Protein Sequences, click ProtSequence under DataClasses.
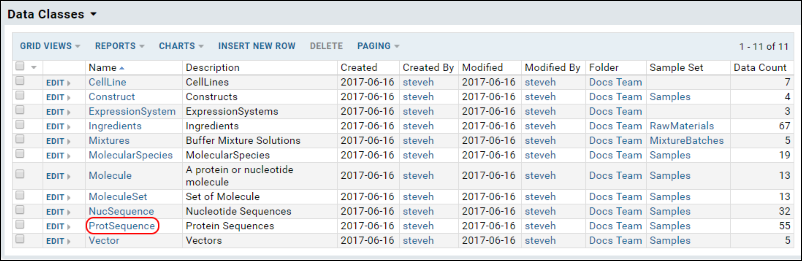
On the ProtSequence DataClass select (Grid Views) > Customize Views.

To add a field, select it on the Available Fields panel. For example, below the Mass field is being added.

To remove a field, click that field's X in the Selected Fields panel. For example, below the Avg. Mass field is being removed.

To change a field name, click that field's Cog in the Selected Fields panel. For example, below the field Organism is being changed to Species.

When the changes have been made as desired, click Save in the lower right.
Save the grid as the Default grid view for this page.

To change field order, drag-and-drop the fields in the Selected Fields panel.
Once the changes have been saved, they will be reflected in the Biologics application.

Details Pages
To control the fields on a details page, follow the same process as described above, except save the view using the name "BIOLOGICSDETAILS", capitalized and one word.

Once saved, the corresponding details view will reflect the changed UI.

Entry Forms
To modify an entity's entry form, modify the fields for that entity. For example, to add a field "NIH Registry Number" to the Cell Line entity:
- In the Data Classes list, click CellLine.
- In the Data Class Properties panel, click Edit Fields.
- Click Add Field, and specify its Name, Label, and Data Type.

- Click Save.
- The field will appear in the entry form for Cell Lines.

Follow similar procedures to delete or modify entry form fields.
Sample Management
Samples and Sample Sets Within LabKey Server
Samples Sets are used to group together Samples and apply shared properties to them.
Sample Set Documentation
Sample Sets and Detail Pages
- Related Topic: Configuring Grids, Detail Views, and Entry Forms
The Samples dashboard shows the sets of samples available, and the relative volume for each in each month. This topic covers working with registered sample data. Click the Samples tile from the home page, or click Samples in the menu bar from within the application.
To learn various methods for generating new sample IDs, see Generate SamplesView Sample Sets and Details
Sample sets listed on the left (or accessable from the Samples menu bar) can be viewed as a datagrid for further sorting or filtering. Click a cell in the grid to see samples for that sample set in that specific month.

Click the sample set name link (to the left of the table or from the Samples menu in the menu bar to see a grid view of that sample set across all months.
Edit Some Details
From any Samples grid view, click a name to see details of that sample, and edit certain fields.
Fields that are used internally, such as the name, folder location, containing sample set, may not be edited, but descriptions and other low-risk fields can be changed here. Changes made here are audited.
Related Topics
Generate Samples
To generate new samples within LabKey Biologics, you can directly upload using LabKey server, or use the Sample Generation wizard described in this topic.
Open the Wizard
There are three entry points for the sample generation wizard:
- From an entity or sample detail page. Click Create Samples.
- From a grid of entities or samples. Having selected one or more rows in the grid, click Create Samples. This allows you to create samples with the selected entites or samples as parents.
- From a grid of samples. With none selected, click Create Samples. This allows you to create samples initially without parents, although you can edit this.
Wizard Fields
The only fields required for generating Sample IDs are:
- Target Sample Set: selected or prepopulated via one of the methods for entering the wizard
- Quantity
Parents
If desired, one or more parents of the new sample can be specified. Click Add Parent, then select the type of parent to be added from the list of Data Classes and Sample Sets available. More than one type of parent may be selected if needed, and there is no requirement that each new sample have parents from every type selected.
If you are generating new sample IDs from an existing sample or entity, it will already be prepopulated as a parent, with the type of entity preselected as a parent type.
In each sample group section, you will then see a selection pulldown for each type of parent selected above. Use the selection pulldowns to select the desired parents from the members of the set or class selected above. Remember you can type ahead to filter the drop down menus.
Sample Groups
At least one sample group is required, and created by default for the quantity given, but more may be added using the Add Sample Group button. Each sample group can be populated with a different quantity and descriptions. The parent selection process works as above, with each sample group having a unique set of parents.
Generating Samples
Once the fields in the wizard are completed, click Finish to generate the new samples.
Each sample group will report success or failure to create the samples in that group. If one group fails while another succeeds, the successful group of samples will be created and that group will be disabled within the wizard. You can make changes to the group that raised the error and resubmit until all groups succeed.
Upon successful submission, you will see the sample set filtered to show only your newly created samples.
Assay Management
Set Up Assays
- LabKey Server Docs: Design a New Assay
Assay data in captured in LabKey Biologics by "Assay Designs". Each Assay Design is essentially a table, where each field has a name and data type, for example, the following Assay Design captures data from a Titration experiment.
| SampleID | Expression System | SampleDate | InjVol | Vial | Cal CurveID | Dilution | ResultID | MAb |
|---|---|---|---|---|---|---|---|---|
| 2016_01_22-C3-12-B9-01 | ES-1 | 2016-01-23 | 30 | 2:A,19 | 46188 | 1.0000 | 47836 | 0.2000 |
| 2016_04_21-C7-73-B6-02 | ES-1 | 2016-04-22 | 30 | 2:B,19 | 46188 | 1.0000 | 47835 | 0.2300 |
| 2015_07_12-C5-39-B4-02 | ES-1 | 2015-07-13 | 30 | 2:C,19 | 46188 | 1.0000 | 47834 | 0.3000 |
Assays Requirements
For an Assay Design to work with LabKey Biologics, it must meet the following requirements:
- The key field must be named "SpecimenID".
- The key field must of data type "lookup"
- The key field lookup must point to a Sample Set (or the exp.Materials table).
Create a New Assay
To create a new assay in LabKey Biologics, do the following:
- From the Assays page, click New Assay.
- Select the assay type (this is like a template for your design). For most cases, select General and click Next.
- In the General Assay Designer enter:
- Name field - this should reflect something about the nature of the assay and what is primarily being measured, for example, "Titer", "Immune Score", or "Cell Properties".
- Scroll down to the Data Fields area.
- Click Add Field button to add new fields. Provide their Name, Label, and Type (= the data type).
- Keep adding fields until you data can be adequately captured.
SpecimenID Field = A Lookup
The SpecimenID field get special treatment in LabKey Biologics -- it points to the originating sample or material that is being measured by the assay. The steps below establish a link between the Assay Design and the originating sample/material.
- For the SpecimenID field (already present by default in any General Assay Design), in the Type field, click the dropdown chevron.
- This brings up the Choose Field Type popup menu. Select lookup. (This is a linking data type. "Lookup" means that the SpecimenID will lookup its values in another table, in this case a Sample Set.)
- For Schema select samples.
- For Table select the appropriate Sample Set. In most cases, choose the (String) version of the table. The choice between (Integer) or (String) reflects the primary key of the underlying table. (ExpressionSystemSamples is shown below.)

- Click Apply to confirm the lookup data type.
Finish the Assay Design
- When the fields are sufficiently defined, click Save and Close.
Import Data to the Assay Design
- Once you import data to the assay design, it will be shown in LabKey Biologics.
Related Topics
Media Registration
Registering Ingredients and Raw Materials
Definitions
- Ingredients are virtual entities in LabKey Biologics, and capture the fixed natural properties of a substance. For example, the Ingredient Sodium Chloride includes its molecular weight, melting and boiling points, general description, etc. You register the Ingredient Sodium Chloride only once.
- Raw Materials are the particular physical instantiations of an Ingredient as real "samples" or "bottles". You register multiple bottles of the Raw Material Sodium Chloride, each with different amounts, sources, lot numbers, locations, vessels, etc.
This topic describes viewing registered media and steps for registration of ingredients and materials. The processes for creating mixtures and batches are covered elsewhere.
The Media dashboard provides tiles for registering and managing:
Ingredients
Clicking Ingredients brings you to a grid of available (previously created) ingredients and an Insert New button to create a new ingredient.
Clicking Insert New opens the wizard for creating a new ingredient. This wizard is controlled by the fields in the 'Ingredients' DataClass, which can be customized through LabKey Server by an administrator to include the fields you need. For example, the insert panel for ingredients might look like this:
Once required fields are completed, click Submit and you will see the grid filtered to show only your new ingredient. Click the 'X' on the filter to return to viewing all ingredients.
Raw Materials
The raw materials used in mixtures are listed here.
Click Insert New to add a new raw material. The data entry wizard is similar to that for ingredients; the fields included are determined by the "RawMaterials" data class. For example:
- Ingredient Name (Required)
- Quantity (Required)
- Description
- Manufacturer
- Product Number
- Lot Number
- Volume
- CAS Number
Related Topics
Registering Mixtures (Recipes)
Definitions
- Mixtures are recipes that combine Ingredients using specific preparation steps. Mixtures are virtual entities in LabKey Biologics. Each Mixture is registered only once, but are realized/instantiated multiple times by Batches.
- Batches are realizations of a Mixture recipe. They are physically real formulations produced by following the recipe encoded by some Mixture. Multiple Batches of the same Mixture can be added to the registry, each with its own volume, weight, vessel, location, etc.
Registering Mixtures
The virtual recipes are listed in the Media > Mixtures grid. Find a given Mixture by filtering, sorting, or searching by typing into the "Select" box.
Mixtures are comprised of Ingredients and specific preparation details in a "recipe" that can be registery. There are several ways to reach the mixture creation wizard:
- Create a new mixture "from scratch" using Insert New from Media > Mixtures
- Create a new mixture starting from an existing mixture
- Create a new mixture starting from an existing ingredient
- Registering Mixtures with Unknown Ingredients, Amounts, or Concentrations.
- Registering Batches with Unknowns.
Start a New Mixture "From Scratch"
From the home page or top menu, select Media, then Mixtures and click Insert New above the grid of available mixtures.
Start from an Existing Mixture
Any of the registered mixtures can be clicked for a detailed view. The details view includes general information as well as the mixture’s included ingredients and preparation steps. The details page also includes a Create > Mixture menu in the which opens the create mixture wizard (and includes the current mixture within the wizard).
Start with an Existing Ingredient
Clicking Ingredients brings you to a grid of available (previously created) ingredients and any can be clicked for a detailed view. Select Create > Mixture to open the wizard to create a new mixture which includes the current ingredient within the wizard.

Register a Mixture: Wizard Steps
The new mixture wizard adds a new mixture to the registry, and performs a check to ensure that the mixture name is unique in the registry. Duplicate mixture names are not allowed in the registry.
Wizard Step 1: Details
The Details step asks for basic information about the mixture, including a type such as powder or solution. Once all required fields have been filled in, the Next button will become clickable.

Upon clicking Next, the mixture name is checked against the registry to see if it is already in use. A warning displayed if a duplicate name is found.

Wizard Step 2: Ingredients
The Ingredients step allows you to add single ingredients or existing mixtures to a new mixture recipe, as well as the required amounts and the amount unit. If you started the wizard from an ingredient or mixture it will be prepopulated here.

As part of adding an ingredient, specific what Recipe Measure Type the mixture is using: Mass or Volume. This selection dictates what unit types are available for the ingredients below.
- If Mass is selected, unit types will be: g/kg, g/g, g/mg, g/μg, mol/kg, mol/g, mol/mg, mol/μg
- If Volume is selected, unit types will be: g/kL, g/L, g/mL, g/μL
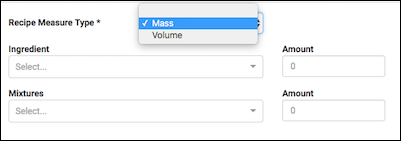
The Ingredient and Mixture select boxes are 'type ahead' searches, that is, as you type, a filtered dropdown of the available options that contain your typed string is shown.

Once an option is selected, a new input will automatically display to allow for more ingredients or mixtures to be added. When a mixture has been selected, more information about the ingredients included and their respective amount/units can be viewed by clicking Show Recipe Ingredients.


When at least one ingredient or mixture and the associated amount/amountUnit fields have been filled in, the Next button becomes enabled. If a selected Ingredient or Mixture is no longer desired, the red minus button on the right can be clicked to remove it.
Wizard Step 3: Preparation
The preparation step lets you add one or more preparation steps with their list of ingredients. The Name and Description inputs will accept any text. When at least one Name and Direction have been included, the Next button will become enabled.

If more preparation steps are desired, click the Add Step button.

Wizard Step 4: Confirmation
The confirmation step summarizes all of the information entered so far. Once submitted, the mixtures grid is shown, filtered to show only the new mixture.

Registering Mixtures with Unknown Ingredients, Amounts, or Concentrations
In cases where you do not know the exact ingredients, amounts, or concentrations of a material, you can still add it to the registry. These scenarios are common when receiving a material from an outside vendor who does not disclose the exact formulation of the product.
In such cases, the mixture registration process is largely the same, except for the Ingredients step, where you can toggle all ingredients and amounts as unknown, or toggle individual amounts as unknown.

Selecting the Ingredients are unknown checkbox disables entry of any further details about the recipe:

Selecting an Unknown amount checkbox disables that row’s amount and unit type inputs:

Note that you can register materials and mixtures without specifying ingredients or amounts, even when you do not explicitly check one of the 'unknown' boxes. But warning messages are provided to confirm that your registration is intentional.

Once registered, mixtures with unknow ingredients and amounts behave just as any other mixture, with a slight change to their detail view to illustrate that an ingredient or concentration may be unknown.


Related Topics
- Registering Ingredients and Raw Materials: Register new ingredients and raw materials.
- Registering Batches
Registering Batches
Batches
Batches represent a specific quantity of a given mixture. The mixture is a virtual entity, i.e. a recipe, used to create the physical batch of material.
To create a new batch, click Insert New while viewing the batches grid, or select Create > Mixture Batch from any mixture detail page to include that mixture in your batch.

The steps in the batch creation wizard are similar to those in the mixture wizard.
Details
On the Details tab, the only required fields is the mixture (i.e., the recipe) to be used to create the batch. Optional fields are: Batch Name, Description, and the Vendor/Part Number. If the Batch Name is not provided, it will be automatically assigned a unique name. Click Next.

Preparation
On the Ingredients tab, each ingredient row is disabled until you add a Desired Batch Yield and Unit. Once selected, the rest of the page will become enabled and populate information
Enter the exact amounts of which raw materials were used to create the mixture recipe. (If ingredients and/or amounts are unknown, see options below.)
For each raw material, specify a source lot by its id number.
Each ingredient in the recipe may have one or more source lots of raw material This is useful when you exhaust one lot and need to use a second lot to complete the batch. For example, using 13mL of Potassium Phosphate empties the bottle, so you need to draw an additional 118mL from a different lot. Specify multiple raw ingredients by clicking Add raw material. (Note that if one or more raw materials are indicated, the option to set an amount or raw material as unknown is no longer available.)

If multiple raw materials were used, the batch details panel will display material lot numbers and the amounts used.

For each preparation step in the mixture recipe, the user can enter any preparation notes necessary in creating this particular batch. Notes are optional, but can provide helpful guidance if something unusual occurred with the batch.
Once all required fields are filled, the Next button will become clickable.

Confirmation
The confirmation step allows the user to view the information they have entered. If necessary, they can click back to return to previous tabs to make any updates. Preparation notes are collapsed, but can be expanded by clicking the right-arrow icon.
Click Finish to register this batch and see the row as entered in the grid.

Registering Batches with Unknowns
When registering batches, the Ingredients step includes the option to mark:
- all materials as unknown
- individual raw materials as unknown
- or individual amounts as unknown
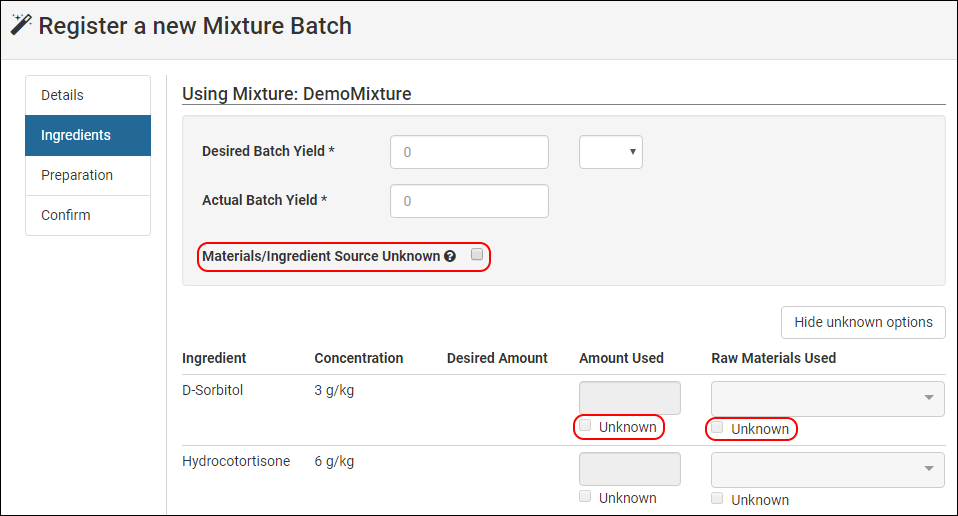
Note the button Show unknown options. When clicked, the UI reveals "Unknown" checkboxes underneath each row's amount and raw material input. Selecting Unknown will then disable the individual input.
To disable all material and amount inputs, select Materials/Ingredient Source Unknown.
Confirmation warnings are provided if the user provides incomplete values and an 'unknown' box is not ticked.

Once entered into the registry, the unknown factors are reflected in the user interface.

Related Topics
Workflow Management
Workflow
[Video: Quick Look: Managing Assay Requests in LabKey Biologics]
Common uses of the Workflow page include:
- Requesting specific work items to be performed, such as running an assay on a given sample.
- Creating "multi-service requests", i.e., umbrella requests with multiple subtasks underneath.
- Assigning work items to a specific user, or chain of users, for fulfillment.
- Tracking job status (incomplete, complete, results invalid, etc.)
- Easy navigation between requests and any associated samples or data results.
The Workflow page supports different types of work requests (with more types available in future releases), for example: general work items, assay run requests, and sample creation requests.
Requests can be created as single stand-alone requests, or as multi-service requests which tie together a group of subtasks.
The Workflow page displays the following panels:
- A histogram showing current requests broken down by type.
- Tabs showing requests (1) opened by and (2) assigned to the currently logged in user.
- Data grids listing the current work items, assay requests, and sample requests.
Create a Work Request
To create a new work request:
- Click Request Work in the upper right corner.

- Fill out details using the entry form.
-
- Title - This title will appear in data grids showing current requests.
- Description - The description should describe the work to be done and the conditions for its completion. (In terms of the LabKey issue tracker, this is the "body" of the request.)
- Assigned To - Assign the task to the person responsible for completing it. Work items may be re-assigned if a more appropriate person is found, or if a series of people are required to complete the task.
- Priority - Assign an number between 0 and 4 to indicate the importance of the task. 0 is most important, 4 is least important.
- Notify List - Add one or more users who will be notified via email each time the work item is updated.
- Related Samples and Entities - Click the green plus sign to add one or more related entites/samples, for example, samples to be consumed in an assay run. Use the dropdowns to select the Type and ID of each item. The dropdowns also function as search boxes: enter text to filter down the options.
- List of subtasks - If the work item consists of multiple tasks, click the green plus sign to add one or more subtasks, such as Assay run requests, sample requests, or any task type that has been configured. Note that subtask names are generated by concatenating: parent request name + the subtask type. For each subtask use the dropdowns to specify:
-
- Type of Service - Specify an assay run service or a sample creation service.
- Service - For assays, this field indicates which sort of assay to run. For samples, this field indicates the type of sample to create.
- Assigned To - Assign the task to the person responsible for completing it. Work items may be re-assigned if a more appropriate person is found, or if a series of people are required to complete the task.
- Description: Optional field for providing details and relevant information.
- Attachments - To add file attachments to the work request, click the Paperclip icon or drag-and-drop the files from your operating system.
- Click Create to submit the work request. Users referred to in the entry form will be notified of the new request(s) via email.
- Submitted requests are displayed in the data grids at the bottom of the page. Note that subtasks are prepended with the name of the parent request.

- The details view for a parent request also shows the related subtasks.

Processing Requests
From a user's perspective, the request tracker functions much like the LabKey issue tracker. A typical workflow looks like the following:
- A user begins the process by creating a request for assays to be run and assigning the request to some appropriate person, such as a lab technician, for fulfillment.
- Once the task is complete (for example, the assay has been run and the data is available) the request is "resolved" and assigned back to the original requester.
- If the original requester is satisfied that the work is complete, then the request is "closed".
The assay tracker also provides for adverse events in the workflow. For example, duplicate requests for assay runs can be resolved as "Duplicate", or requests can be resolved as "Rejected" if for some reason the request cannot be completed. The available resolution states are not fixed, and be adjusted by an administrator (see the Administrator docs for documentation).
Related Topics
- Assay Request Tracker: Administrator Documentation - Explains how to customize the system, such as populating the dropdowns with custom values.
- Issue Tracker: Administration - Explains how to administer the issue tracking system generally, such as controlling the "Assigned To" dropdown, adding custom fields, etc.
- Service Request Tracker Set Up - Explain how to set up subtasks in work requests.
Service Request Tracker Set Up
1. Create an issue tracker or use a pre-existing one. This will track the subtasks and cannot be the same as the parent issue tracker. Go to (Admin) > Go To Module > Issues and click Add New Row. For detailed docs see Issue Tracker: Administration.
2. Set up a column on the service tracker as a lookup to what your services are. You can create a new column, or use an existing column. For example, the image below shows how to create a lookup to the exp.SampleSets table. This will convert the column into a dropdown populated with sample sets to choose from.
In the case of Assay requests, the column is already provided as the "Assay" column, a preconfigured column to the assay.AssayList table.
3. Go to the Schema Browser ( (Admin) > Developer Links > Schema Browser) and edit the metadata for the service tracker.
4. On the Edit Metadata page click Edit Source at the bottom.
5. You should see a blank text area for editing "XML Metadata". Replace <TRACKER TABLE NAME> and <TRACKER SERVICE COLUMN> with the name of the service tracker table and the name of the service column you setup in step 2.
<tables xmlns="http://labkey.org/data/xml">
<table tableName="<TRACKER TABLE NAME>" tableDbType="NOT_IN_DB">
<columns>
<column columnName="<TRACKER SERVICE COLUMN>">
<conceptURI>biologics-workflow-service</conceptURI>
</column>
</columns>
</table>
</tables>
6. Click Save and Finish to complete the configuration.
7. Now you can create new service requests in Biologics.
