Table of Contents |
guest 2025-07-16 |
Tutorial: Hello World Module
Map of Module Files
Example Modules
Modules: Queries, Views and Reports
Module Directories Setup
Module Query Views
Module SQL Queries
Module R Reports
Module HTML and Web Parts
Modules: JavaScript Libraries
Modules: Assay Types
Tutorial: Define an Assay Type in a Module
Assay Custom Domains
Assay Custom Views
Example Assay JavaScript Objects
Assay Query Metadata
Customize Batch Save Behavior
SQL Scripts for Module-Based Assays
Transformation Scripts
Example Workflow: Develop a Transformation Script (perl)
Example Transformation Scripts (perl)
Transformation Scripts in R
Transformation Scripts in Java
Transformation Scripts for Module-based Assays
Run Properties Reference
Transformation Script Substitution Syntax
Warnings in Tranformation Scripts
Modules: ETLs
Tutorial: Extract-Transform-Load (ETL)
ETL Tutorial: Set Up
ETL Tutorial: Run an ETL Process
ETL Tutorial: Create a New ETL Process
ETL: User Interface
ETL: Configuration and Schedules
ETL: Column Mapping
ETL: Queuing ETL Processes
ETL: Stored Procedures
ETL: Stored Procedures in MS SQL Server
ETL: Functions in PostgreSQL
ETL: Check For Work From a Stored Procedure
ETL: SQL Scripts
ETL: Remote Connections
ETL: Logs and Error Handling
ETL: All Jobs History
ETL: Examples
ETL: Reference
Modules: Java
Module Architecture
Getting Started with the Demo Module
Creating a New Java Module
The LabKey Server Container
Implementing Actions and Views
Implementing API Actions
Integrating with the Pipeline Module
Integrating with the Experiment Module
Using SQL in Java Modules
GWT Integration
GWT Remote Services
Java Testing Tips
HotSwapping Java classes
Deprecated Components
Modules: Folder Types
Modules: Query Metadata
Modules: Report Metadata
Modules: Custom Footer
Modules: SQL Scripts
Modules: Database Transition Scripts
Modules: Domain Templates
Deploy Modules to a Production Server
Upgrade Modules
Main Credits Page
Module Properties Reference
Develop Modules
A wide variety of resources can be used, including, R reports, SQL queries and scripts, API-driven HTML pages, CSS, JavaScript, images, custom web parts, XML assay definitions, and compiled Java code. Much module development can be accomplished without compiling Java code, letting you directly deploy and test module source, oftentimes without restarting the server.
Module Functionality
| Functionalityoooooooo | Description | Docsooooooooooooooooooooooooooo |
|---|---|---|
| Queries, Views, and Reports | A module that includes queries, reports, and/or views directories. Create file-based SQL queries, reports, views, web parts, and HTML/JavaScript client-side applications. No Java code required, though you can easily evolve your work into a Java module if needed. | Modules: Queries, Views and Reports |
| Assay | A module with an assay directory included, for defining a new assay type. | Modules: Assay Types |
| Extract-Transform-Load | A module with an etl directory included, for configuring data transfer and synchronization between databases. | Modules: ETLs |
| Script Pipeline | A module with a pipeline directory included, for running scripts in sequence, including R scripts, JavaScript, Perl, Python, etc. | Script Pipeline: Running R and Other Scripts in Sequence |
| Java | A module with a Java src directory included. Develop Java-based applications to create server-side code. | Modules: Java |
Do I Need to Compile Modules?
Modules do not need to be compiled, unless they contain Java code. Most module functionality can be accomplished without the need for Java code, including "CRUD" applications (Create-Retrieve-Update-Delete applications) that provide views and reports on data on the server, and provide some way for users to interact with the data. These applications will typically use some combination of the following client APIs: LABKEY.Query.selectRows, insertRows, updateRows, and deleteRows.
Also note that client-side APIs are generally guaranteed to be stable, while server-side APIs are not guaranteed to be stable and are liable to change as the LabKey Server code base evolves -- so modules based on the server API may require changes to keep them up to date.
More advanced client functionality, such as defining new assay types, working with the security API, and manipulating studies, can also be accomplished with a simple module without Java.
To create your own server actions (i.e., code that runs on the server, not in the client), Java is generally required. Trigger scripts, which run on the server, are are an exception: trigger scripts are a powerful feature, sufficient in many cases to avoid the need for Java code. Note that Java modules require a build/compile step, but modules without Java code don't need to be compiled before deployment to the server.
Module Development Setup
Use the following topic to set up a development machine for building LabKey modules: Set up a Development Machine
Topics
The topics below show you how to create a module, how to develop the various resources within the module, and how to package and deploy it to LabKey Server.
- Map of Module Files - An overview of the features and file types you can include in modules.
- Example Modules - Representative example modules from the LabKey Server open source project.
- Modules: Queries, Views and Reports - Build a more complex file-based module.
- Modules: Assay Types - Module-based custom assay types, to base assay designs on.
- Modules: ETLs - Module-based Extract-Transform-Load functionality.
- Modules: Java - Including Java in modules.
- Modules: Folder Types - Module-based custom folder types.
- Modules: SQL Scripts - Running SQL scripts on module load.
- Deploy Modules to a Production Server - Deploying modules for production.
- Upgrade Modules - How to handle module upgrades.
- MiniProfiler - Developer tool shows which queries were run on a page, and performance information for each query.
- Main Credits Page - Include a credits page.
- Module Properties Reference - Reference for module.properties/module.xml files.
Tutorial: Hello World Module
LabKey Server's functionality is packaged inside of modules. For example, the query module handles the communication with the databases, the wiki module renders Wiki/HTML pages in the browser, the assay module captures and manages assay data, etc.
You can extend the functionality of the server by adding your own module. Here is a partial list of things you can do with a module:
- Create a new assay type to capture data from a new instrument.
- Add a new set of tables and relationships (= a schema) to the database by running a SQL script.
- Develop file-based SQL queries, R reports, and HTML views.
- Build a sequence of scripts that process data and finally insert it into the database.
- Define novel folder types and web part layouts.
- Set up Extract-Transform-Load (ETL) processes to move data between databases.
The following tutorial shows you how to create your own "Hello World" module and deploy it to a local testing/development server.
Set Up a Development Machine
In this step you will set up a test/development machine, which compiles LabKey Server from its source code.
If you already have a working build of the server, you can skip this step.
- If necessary, uninstall any instances of LabKey Server that were installed using the Windows Graphical Installer, as an installer-based server and a source-based server cannot run together simultaneously on the same machine. Use the Windows uninstaller at: Control Panel > Uninstall a program. If you see LabKey Server in the list of programs, uninstall it.
- Download the server source code and complete an initial build of the server by completing the steps in the following topic: Set up a Development Machine
- Before you proceed, build and deploy the server. Confirm that the server is running by visiting the URL http://localhost:8080/labkey/project/home/begin.view?
- For the purposes of this tutorial, we will call the location where you have synced the server source code LABKEY_SRC. On Windows, a typical location for LABKEY_SRC would be C:/dev/trunk
Module Properties
In this step you create the main directory for your module and set basic module properties.
- Go to LABKEY_SRC, the directory where you synced to the server source code, and locate the directory externalModules.
- Inside LABKEY_SRC/externalModules, create a directory named "helloworld".
- Inside the helloworld directory, create a file named "module.properties".
- Add the following property/value pairs to module.properties. This is a minimal list of properties needed for deployment and testing. You can add a more complete list of properties later on, including your name, links to documentation, required server and database versions, etc. For a complete list of available properties see Module Properties Reference.
Name: HelloWorld
ModuleClass: org.labkey.api.module.SimpleModule
Version: 1.0
Build and Deploy the Module
- Open the file LABKEY_SRC/server/standard.modules (This file controls which modules are included in the build.)
- Add this line to the file:
externalModules/helloworld
- Build the server.
- Open a command window.
- Go to directory LABKEY_SRC/server
- Call the ant task:
ant build
- Start the server, either in IntelliJ by click the "Debug" button, or by running the Tomcat startup script appropriate for your operating system (located in TOMCAT_HOME/bin).
Confirm the Module Has Been Deployed
- In a browser go to: http://localhost:8080/labkey/project/home/begin.view?
- Sign in.
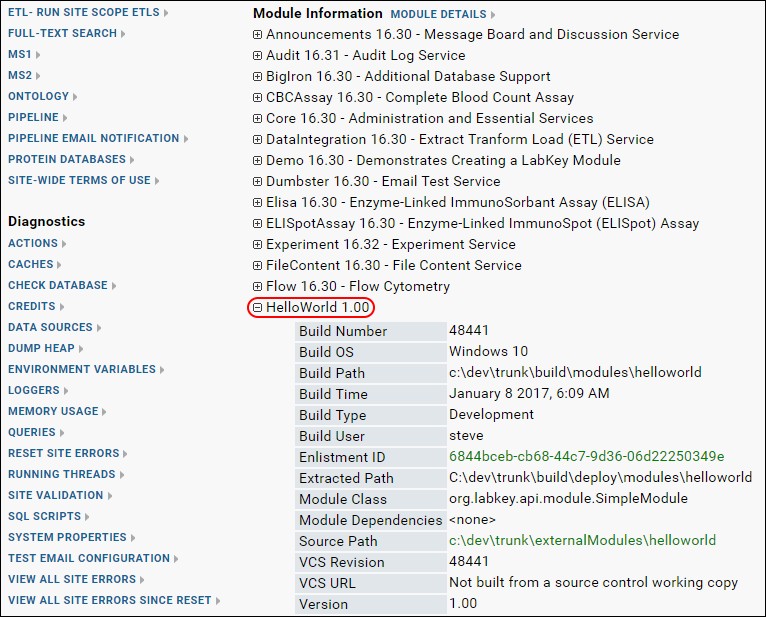
- Confirm that HelloWorld has been deployed to the server by going to Admin > Site > Admin Console. Scroll down to Module Information (in the right hand column). Open the node HelloWorld. Notice the module properties you specified are displayed here: Name: HelloWorld, Version: 1.0, etc.
Add a Default Page
Each module has a default home page called "begin.view". In this step we will add this page to our module. The server interprets your module resources based on a fixed directory structure. By reading the directory structure and the files inside, the server knows their intended functionality. For example, if the module contains a directory named "assays", this tells the server to look for XML files that define a new assay type. Below, we will create a "views" directory, telling the server to look for HTML and XML files that define new pages and web parts.
- Inside helloworld, create a directory named "resources".
- Inside resources, create a directory named "views".
- Inside views, create a file named "begin.html". (This is the default page for any module.)
helloworld
│ module.properties
└───resources
└───views
begin.html
- Open begin.html in a text editor, and add the following HTML code:
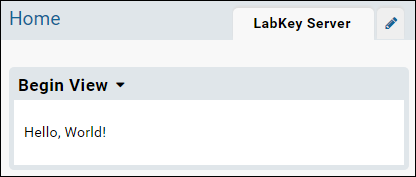
<p>Hello, World!</p>
Test the Module
- Build the server by calling 'ant build'.
- Wait for the server to redeploy.
- Enable the module in some test folder:
- Navigate to some test folder on your server.
- Go to Admin > Folder > Management and click the Folder Type tab.
- In the list of modules on the right, place a checkmark next to HelloWorld.
- Click Update Folder.
- Confirm that the view has been deployed to the server by going to Admin > Go to Module > HelloWorld.
- The following view will be displayed:

Modify the View with Metadata
You can control how a view is displayed by using a metadata file. For example, you can define the title, framing, and required permissions.
- Add a file to the views directory named "begin.view.xml". Note that this file has the same name (minus the file extension) as begin.html: this tells the server to apply the metadata in begin.view.xml to being.html.
helloworld
│ module.properties
└───resources
└───views
begin.html
begin.view.xml
- Add the following XML to begin.view.xml. This tells the server to: display the title 'Begin View', display the HTML without any framing, and that Reader permission is required to view it.
<view xmlns="http://labkey.org/data/xml/view"
title="Begin View"
frame="none">
<permissions>
<permission name="read"/>
</permissions>
</view>
- Refresh your browser to see the result. (You do not need to rebuild or restart the server.)
- The begin view now looks like the following:

- Experiment with other possible values for the 'frame' attribute:
- portal (If no value is provided, the default is 'portal'.)
- title
- dialog
- div
- left_navigation
- none
- When you are ready to move to the next step, set the 'frame' attribute back to 'portal'.
Hello World Web Part
You can also package the view as a web part using another metadata file.
- In the helloworld/resources/views directory add a file named "begin.webpart.xml". This tells the server to surface the view inside a webpart. Your module now has the following structure:
helloworld
│ module.properties
└───resources
└───views
begin.html
begin.view.xml
begin.webpart.xml
- Paste the following XML into begin.webpart.xml:
<webpart xmlns="http://labkey.org/data/xml/webpart"
title="Hello World Web Part">
<view name="begin"/>
</webpart>
- Return to your test folder using the hover menu in the upper left.
- In your test folder, click the dropdown <Select Web Part>.
- Select the web part Hello World Web Part and click Add.
- The following web part will be added to the page:
Hello User View
The final step provides more interesting view which uses the JavaScript API to retrieve information about the current users.
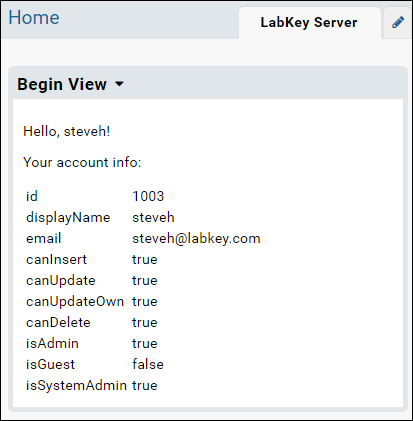
- Open begin.html and replace the HTML with the following.
- Refresh the browser to see the changes. (You can directly edit the file begin.html in the module -- the server will pick up the changes without needing to rebuild or restart.)
<p>Hello, <script>
document.write(LABKEY.Security.currentUser.displayName);
</script>!</p>
<p>Your account info: </p>
<table>
<tr><td>id</td><td><script>document.write(LABKEY.Security.currentUser.id); </script><td></tr>
<tr><td>displayName</td><td><script>document.write(LABKEY.Security.currentUser.displayName); </script><td></tr>
<tr><td>email</td><td><script>document.write(LABKEY.Security.currentUser.email); </script><td></tr>
<tr><td>canInsert</td><td><script>document.write(LABKEY.Security.currentUser.canInsert); </script><td></tr>
<tr><td>canUpdate</td><td><script>document.write(LABKEY.Security.currentUser.canUpdate); </script><td></tr>
<tr><td>canUpdateOwn</td><td><script>document.write(LABKEY.Security.currentUser.canUpdateOwn); </script><td></tr>
<tr><td>canDelete</td><td><script>document.write(LABKEY.Security.currentUser.canDelete); </script><td></tr>
<tr><td>isAdmin</td><td><script>document.write(LABKEY.Security.currentUser.isAdmin); </script><td></tr>
<tr><td>isGuest</td><td><script>document.write(LABKEY.Security.currentUser.isGuest); </script><td></tr>
<tr><td>isSystemAdmin</td><td><script>document.write(LABKEY.Security.currentUser.isSystemAdmin); </script><td></tr>
</table>
- Once you've refreshed the browser, the web part will display the following.
Make a .module File
You can distribute and deploy a module to production server by making a helloworld.module file (a renamed .zip file).
- In anticipation of deploying the module on a production server, add the property 'BuildType: Production' to the module.properties file:
Name: HelloWorld
ModuleClass: org.labkey.api.module.SimpleModule
Version: 1.0
BuildType: Production
- Then build the module:
ant build
- The build process creates a helloworld.module file at:
LABKEY_SRC/build/deploy/modules/helloworld.module
This file can be deployed by copying it to another server's externalModule directory. When the server detects changes in this directory, it will automatically unzip the .module file and deploy it. You may need to restart the server to fully deploy the module.
Related Topics
These tutorials show more functionality that you can package as a module:
Map of Module Files
Module Directories and Files
The following directory structure follows the pattern for modules as they are checked into source control. The structure of the module as deployed to the server is somewhat different, for details see below and the topic Module Properties Reference. If your module contains Java code or Java Server Pages (JSPs), you will need to compile it before it can be deployed.
Items shown in lowercase are literal values that should be preserved in the directory structure; items shown in UPPERCASE should be replaced with values that reflect the nature of your project.
MODULE_NAME │ module.properties docs │ module.xml docs, example └──resources ├───assay docs ├───config │ module.xml docs, example ├───credits docs, example ├───domain-templates docs ├───etls docs ├───folderTypes docs ├───olap example ├───pipeline docs, example ├───queries docs │ └───SCHEMA_NAME │ │ QUERY_NAME.js docs, example │ │ QUERY_NAME.query.xml docs, example │ │ QUERY_NAME.sql example │ └───QUERY_NAME │ VIEW_NAME.qview.xml docs, example ├───reports docs │ └───schemas │ └───SCHEMA_NAME │ └───QUERY_NAME │ MyRScript.r example │ MyRScript.report.xml docs, example │ MyRScript.rhtml docs │ MyRScript.rmd docs ├───schemas docs │ │ SCHEMA_NAME.xml example │ └───dbscripts │ ├───postgresql │ │ SCHEMA_NAME-X.XX-Y.YY.sql example │ └───sqlserver │ SCHEMA_NAME-X.XX-Y.YY.sql example ├───scripts docs, example ├───views docs │ VIEW_NAME.html example │ VIEW_NAME.view.xml example │ TITLE.webpart.xml example └───web docs └───MODULE_NAME SomeImage.jpg somelib.lib.xml SomeScript.js example
Module Layout - As Source
If you are developing your module inside the LabKey Server source, use the following layout. The standard build targets will automatically assemble the directories for deployment. In particular, the standard build target makes the following changes to the module layout:
- Moves the contents of /resources one level up into /mymodule.
- Uses module.properties to create the file config/module.xml via string replacement into an XML template file.
- Compiles the Java /src dir into the /lib directory.
mymodule
├───module.properties
├───resources
│ ├───assay
│ ├───etls
│ ├───folderTypes
│ ├───queries
│ ├───reports
│ ├───schemas
│ ├───views
│ └───web
└───src (for modules with Java code)
Module Layout - As Deployed
The standard build targets transform the source directory structure above into the form below for deployment to Tomcat.
mymodule
├───assay
├───config
│ └───module.xml
├───etls
├───folderTypes
├───lib (holds compiled Java code)
├───queries
├───reports
├───schemas
├───views
└───web
Related Topics
Example Modules
To acquire the source code for these modules, enlist in the LabKey Server open source project: Enlisting in the Version Control Project
| Module Location | Description / Highlights |
|---|---|
| server/customModules | This directory contains numerous client modules, in most cases Java modules. |
| server/modules | The core modules for LabKey Server are located here, containing the core server action code (written in Java). |
| server/test | The test module runs basic tests on the server. Contains many basic examples to clone from. |
| externalModules | Other client modules. |
Other Resources
- Creating a New Java Module - Using an Ant target, create a template Java module that you can build out from.
- Getting Started with the Demo Module - A sample module demonstrates Java module development.
- Trigger Scripts - Module-based trigger scripts, schemas created by SQL scripts.
- Community Modules - Descriptions of the modules in the standard distribution of LabKey Server.
Modules: Queries, Views and Reports
The Scenario
Suppose that you want to present a series of R reports, database queries, and HTML views. The end-goal is to deliver these to a client as a unit that can be easily added to their existing LabKey Server installation. Once added, end-users should not be able to modify the queries or reports, ensuring that they keep running as expected. The steps below show how to fulfill these requirements using a file-based module.
Steps:
- Module Directories Setup
- Module Query Views
- Module SQL Queries
- Module R Reports
- Module HTML and Web Parts
Use the Module on a Production Server
This tutorial is designed for developers who build LabKey Server from source. But even if you are not a developer and do not build the server from source, you can get a sense of how modules by work by installing the module that is the final product of this tutorial. To install the module, download reportDemo.module and copy the file into the directory LABKEY_HOME\externalModules (on a Windows machine this directory is typically located at C:\Program Files(x86)\LabKey Server\externalModules). Notice that the server will detect the .module file and unzip it, creating a directory called reportDemo, which is deployed to the server. Look inside reportDemo to see the resources that have been deployed to the server. Read through the steps of the tutorial to see how these resources are surfaced in the user interface.
First Step
Module Directories Setup
- queries - Holds SQL queries and views.
- reports - Holds R reports.
- views - Holds user interface files.
Set Up a Dev Machine
Complete the topics below. This will set up a machine that can build LabKey Server (and the proteomics tools) from source.
Install Sample Data
- Install the Proteomics sample data by following the instructions in these topics:
Create Directories
- Go to the externalModules/ directory, and create the following directory structure and standard.modules file:
reportDemo
│ module.properties
└───resources
├───queries
├───reports
└───views
Add the following contents to module.properties:
Module Class: org.labkey.api.module.SimpleModule
Name: ReportDemo
Build the Module
- Open the file module.properties and add the following line:
externalModules/reportDemo
- In a command shell, go to the 'server' directory, for example, 'cd C:\dev\labkey-src\trunk\server'.
- Call 'ant build' to build the module.
- Restart the server to deploy the module.
Enable Your Module in a Folder
To use a module, enable it in a folder.
- Go to the LabKey Server folder where you want add the module functionality.
- Select Admin -> Folder -> Management -> Folder Type tab.
- Under the list of Modules click on the check box next to ReportDemo to activate it in the current folder.
Start Over | Next Step
Module Query Views
- SQL queries on the database (.SQL files)
- Metadata on the above queries (.query.xml files).
- Named views on pre-existing queries (.qview.xml files)
- Trigger scripts attached to a query (.js files) - these scripts are run whenever there an event (insert, update, etc.) on the underlying table.
Additionally, if you wish to just create a default view that overrides the system generated one, be sure to just name the file as .qview.xml, so there is no actual name of the file. If you use default.qview.xml, this will create another view called "default", but it will not override the existing default.
Create an XML-based SQL Query
- Add two directories (ms2 and Peptides) and a file (High Prob Matches.qview.xml), as shown below.
- The directory structure tells LabKey Server that the view is in the "ms2" schema and on the "Peptides" table.
reportDemo │ module.properties └───resources ├───queries │ └───ms2 │ └───Peptides │ High Prob Matches.qview.xml │ ├───reports └───views
View Source
The view will display peptides with high Peptide Prophet scores (greater than or equal to 0.9).
- Save High Prob Matches.qview.xml with the following content:
<customView xmlns="http://labkey.org/data/xml/queryCustomView">
<columns>
<column name="Scan"/>
<column name="Charge"/>
<column name="PeptideProphet"/>
<column name="Fraction/FractionName"/>
</columns>
<filters>
<filter column="PeptideProphet" operator="gte" value="0.9"/>
</filters>
<sorts>
<sort column="PeptideProphet" descending="true"/>
</sorts>
</customView>
- The root element of the qview.xml file must be <customView> and you should use the namespace indicated.
- <columns> specifies which columns are displayed. Lookups columns can be included (e.g., "Fraction/FractionName").
- <filters> may contain any number of filter definitions. (In this example, we filter for rows where PeptideProphet >= 0.9). (docs: <filter>)
- <sorts> section will be applied in the order they appear in this section. In this example, we sort descending by the PeptideProphet column. To sort ascending simply omit the descending attribute.
See the View
To see the view on the ms2.Peptides table:
- Build and restart the server.
- Go to the Peptides table and click Grid Views -- the view High Prob Matches has been added to the list. (Admin > Developer Links > Schema Browser. Open ms2, scroll down to Peptides. Select Grid Views > High Prob Matches.)
Previous Step | Next Step
Module SQL Queries
If supplied, the metadata file should have the same name as the .sql file, but with a ".query.xml" extends (e.g., PeptideCounts.query.xml) (docs: query.xsd)
Below we will create two SQL queries in the ms2 schema.
- Add two .sql files in the queries/ms2 directory, as follows:
reportDemo │ module.properties └───resources ├───queries │ └───ms2 │ │ PeptideCounts.sql │ │ PeptidesWithCounts.sql │ └───Peptides │ High Prob Matches.qview.xml ├───reports └───views
Add the following contents to the files:
PeptideCounts.sql
SELECT
COUNT(Peptides.TrimmedPeptide) AS UniqueCount,
Peptides.Fraction.Run AS Run,
Peptides.TrimmedPeptide
FROM
Peptides
WHERE
Peptides.PeptideProphet >= 0.9
GROUP BY
Peptides.TrimmedPeptide,
Peptides.Fraction.Run
PeptidesWithCounts.sql
SELECT
pc.UniqueCount,
pc.TrimmedPeptide,
pc.Run,
p.PeptideProphet,
p.FractionalDeltaMass
FROM
PeptideCounts pc
INNER JOIN
Peptides p
ON (p.Fraction.Run = pc.Run AND pc.TrimmedPeptide = p.TrimmedPeptide)
WHERE pc.UniqueCount > 1
Note that the .sql files may contain spaces in their names.
See the SQL Queries
- Build and restart the server.
- To view your SQL queries, go to the schema browser at Admin -> Developer Links -> Schema Browser.
- On the left side, open the nodes ms2-> user-defined queries -> PeptideCounts.
Optionally, you can add metadata to these queries to enhance them. See Modules: Query Metadata.
Previous Step | Next Step
Module R Reports
Below we'll make an R report script that is associated with the PeptidesWithCounts query (created in the previous step).
- In the reports/ directory, create the following subdirectories: schemas/ms2/PeptidesWithCounts, and a file named "Histogram.r", as shown below:
- Open the Histogram.r file, enter the following script, and save the file. (Note that .r files may have spaces in their names.)
png(
filename="${imgout:labkeyl_png}",
width=800,
height=300)
hist(
labkey.data$fractionaldeltamass,
breaks=100,
xlab="Fractional Delta Mass",
ylab="Count",
main=NULL,
col = "light blue",
border = "dark blue")
dev.off()
Report Metadata
Optionally, you can add associated metadata about the report. See Modules: Report Metadata.
Test your SQL Query and R Report
- Go to the Query module's home page (Admin -> Go to Module -> Query). Note that the home page of the Query module is the Query Browser.
- Open the ms2 node, and see your two new queries in the user-defined queries section.
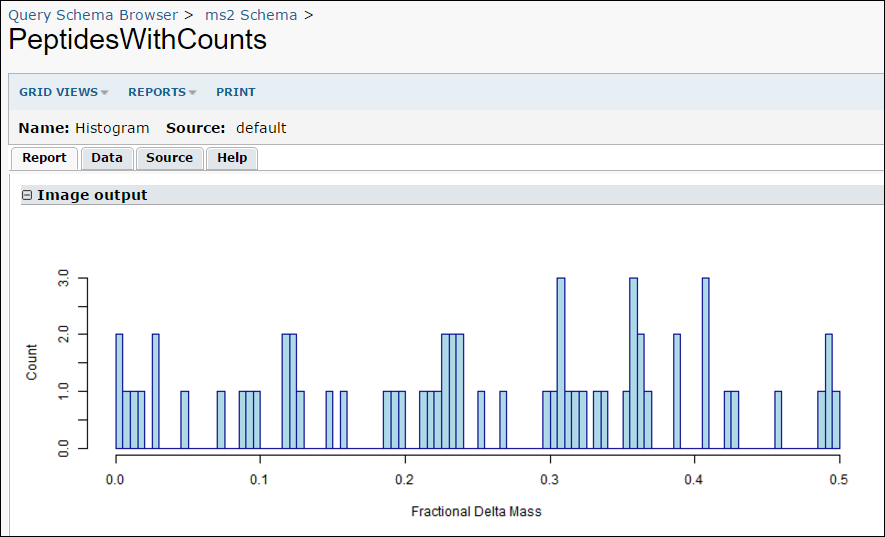
- Click on PeptidesWithCounts and then View Data to run the query and view the results.
- While viewing the results, you can run your R report by selecting Views -> Histogram.
Previous Step | Next Step
Module HTML and Web Parts
Since getting to the Query module's start page is not obvious for most users, we will provide an HTML view for a direct link to the query results. You can do this in a wiki page, but that must be created on the server, and our goal is to provide everything in the module itself. Instead we will create an HTML view and an associated web part.
Add an HTML Page
Under the views/ directory, create a new file named reportdemo.html, and enter the following HTML:
<p>
<a id="pep-report-link"
href="<%=contextPath%><%=containerPath%>/query-executeQuery.view?schemaName=ms2&query.queryName=PeptidesWithCounts">
Peptides With Counts Report</a>
</p>
Note that .html view files must not contain spaces in the file names. The view servlet expects that action names do not contain spaces.
Use contextPath and containerPath
Note the use of the <%=contextPath%> and <%=containerPath%> tokens in the URL's href attribute. These tokens will be replaced with the server's context path and the current container path respectively. For syntax details, see LabKey URLs.
Since the href in this case needs to refer to an action in another controller, we can't use a simple relative URL, as it would refer to another action in the same controller. Instead, use the contextPath token to get back to the web application root, and then build your URL from there.
Note that the containerPath token always begins with a slash, so you don't need to put a slash between the controller name and this token. If you do, it will still work, as the server automatically ignores double-slashes.
Define a View Wrapper
This file has the same base-name as the HTML file, "reportdemo", but with an extension of ".view.xml". In this case, the file should be called reportdemo.view.xml, and it should contain the following:
<view xmlns="http://labkey.org/data/xml/view"
frame="none" title="Report Demo">
</view>
Define a Web Part
To allow this view to be visible inside a web part create our final file, the web part definition. Create a file in the views/ directory called reportdemo.webpart.xml and enter the following content:
<webpart xmlns="http://labkey.org/data/xml/webpart" title="Report Demo">
<view name="reportdemo"/>
</webpart>
After creating this file, you should now be able to refresh the portal page in your folder and see the "Report Demo" web part in the list of available web parts. Add it to the page, and it should display the contents of the reportdemo.html view, which contains links to take users directly to your module-defined queries and reports.
Your directory structure should now look like this:
externalModules/
ReportDemo/
resources/
reports/
schemas/
ms2/
PeptidesWithCounts/
Histogram.r
queries/
ms2/
PeptideCounts.sql
PeptidesWithCounts.sql
Peptides/
High Prob Matches.qview.xml
views/
reportdemo.html
reportdemo.view.xml
reportdemo.webpart.xml
Set Required Permissions
You might also want to require specific permissions to see this view. That is easily added to the reportdemo.view.xml file like this:
<view xmlns="http://labkey.org/data/xml/view" title="Report Demo">
<permissions>
<permission name="read"/>
</permissions>
</view>
You may add other permission elements, and they will all be combined together, requiring all permissions listed. If all you want to do is require that the user is signed in, you can use the value of "login" in the name attribute.
The XSD for this meta-data file is view.xsd in the schemas/ directory of the project. The LabKey XML Schema Reference provides an easy way to navigate the documentation for view.xsd.
Previous Step
Modules: JavaScript Libraries
- Acquire the library .js file want to use.
- In your module resources directory, create a subdirectory named "web".
- Inside "web", create a subdirectory with the same name as your module. For example, if your module is named 'helloworld', create the following directory structure:
- Copy the library .js file into your directory structure. For example, if you wish to use a JQuery library, place the library file as shown below:
- For any HTML pages that use the library, create a .view.xml file, adding a "dependencies" section.
- For example, if you have a page called helloworld.html, then create a file named helloworld.view.xml next to it:
- Finally add the following "dependencies" section to the .view.xml file:
<view xmlns="http://labkey.org/data/xml/view" title="Hello, World!">
<dependencies>
<dependency path="helloworld/jquery-2.2.3.min.js"></dependency>
</dependencies>
</view>
Note: if you declare dependencies explicitly in the .view.xml file, you don't need to use LABKEY.requiresScript on the HTML page.
Remote Dependencies
In some cases, you can declare your dependency using an URL that points directly to the remote library, instead of copying the library file and distributing it with your module:
<dependency path="https://code.jquery.com/jquery-2.2.3.min.js"></dependency>Related Topics
Modules: Assay Types
Topics
- Tutorial: Define an Assay Type in a Module - Get started writing module-based assays.
- Assay Custom Domains - Create a custom result domain.
- Assay Custom Views - Add custom HTML details views.
- Example Assay JavaScript Objects - JavaScript examples for assay modules.
- Assay Query Metadata - Provide additional properties to a query.
- Transformation Scripts - Transform data before updates and inserts using scripts.
- Customize Batch Save Behavior - Customize batch save behavior using Java.
- Suggestions for Extensible Assays - An informative discussion from the LabKey Support Boards
Examples: Module-Based Assays
There are a handful of module-based assays in the LabKey SVN tree. You can find the modules in <LABKEY_ROOT>/server/customModules. Examples include:
- <LABKEY_ROOT>/server/customModules/exampleassay/resources/assay
- <LABKEY_ROOT>/server/customModules/iaviElisa/elisa/assay/elisa
- <LABKEY_ROOT>/server/customModules/idri/resources/assay/particleSize
File Structure
The assay consists of an assay config file, a set of domain descriptions, and view html files. The assay is added to a module by placing it in an assay directory at the top-level of the module. The assay has the following file structure:
The only required part of the assay is the <assay-name> directory. The config.xml, domain files, and view files are all optional.
This diagram shows the relationship between the pages. The details link will only appear if the corresponding details html view is available.
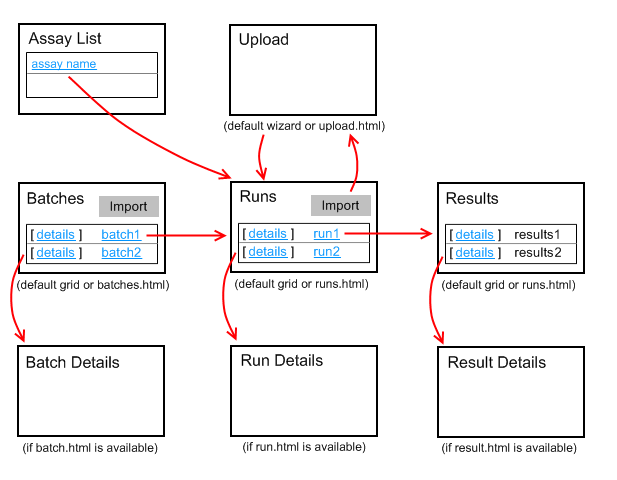
How to Specify an Assay "Begin" Page
Module-based assays can be designed to jump to a "begin" page instead of a "runs" page. If an assay has a begin.html in the assay/<name>/views/ directory, users are directed to this page instead of the runs page when they click on the name of the assay in the assay list.
Tutorial: Define an Assay Type in a Module
To create a module-based assay, you create a set of files that define the new assay design, describe the data import process, and define various types of assay views. The new assay is incorporated into your server when package these files as a module and restart your server. The new type of assay is then available on your server as the basis for new assay designs, in the same way that built-in assay types (e.g., Luminex) are available.
This tutorial explains how to incorporate a ready-made, module-based assay into your LabKey Server and make use of the new type of assay. It does not cover creation of the files that compose a module-based assay. Please refer to the "Related Topics" section below for instructions on how to create such files.
Download
First download a pre-packed .module file and deploy it to LabKey Server.
- Download exampleassay.module. (This is a renamed .zip archive the contains the source files for the assay module.)
Add the Module to your LabKey Server Installation
- On a local build of LabKey Server, copy exampleassay.module to a module deployment directory, such as <LABKEY_HOME>\build\deploy\modules\
- Or
- On a local install of LabKey Server, copy exampleassay.module to this location: <LABKEY_HOME>\externalModules\
- Restart your server. The server will explode the directory.
- Examine the files in the exploded directory. You will see the following structure:
exampleassay
└───assay
└───example
│ config.xml
│
├───domains
│ batch.xml
│ result.xml
│ run.xml
│
└───views
upload.html
- upload.html contains the UI that the user will see when importing data to this type of assay.
- batch.xml, result.xml, and run.xml provide the assay's design, i.e., the names of the fields, their data types, whether they are required fields, etc.
Enable the Module in a Folder
The assay module is now available through the UI. Here we enable the module in a folder.
- Create or select a folder to enable the module in, for example, a subfolder in the Home project.
- Select Admin > Folder > Management and then click the Folder Type tab.
- Place a checkmark next to the exampleassay module (under the "Modules" column on the right).
- Click the Update Folder button.
Use the Module's Assay Design
Next we create a new assay design based the module.
- Select Admin > Manage Assays.
- On the Assay List page, click New Assay Design.
- Select LabKey Example and click Next.
- Name this assay "FileBasedAssay"
- Leave all other fields at default values and click Save and Close.
Import Data to the Assay Design
- Download these two sample assay data files:
- Click on the new FileBasedAssay in the Assay List.
- Click the Import Data button.
- Note that instead of the standard UI for importing data, you will see the custom UI for importing data defined by upload.html in exampleassay\assay\example\views.
- Also note the URL to the custom upload page. It follows the pattern: http://myserver.org/labkey/myproject/myfolder/assay-moduleAssayUpload.view?rowId=1
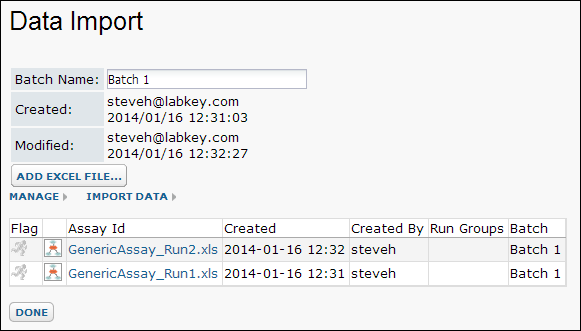
- Enter a value for Batch Name, for example, "Batch 1"
- Click Add Excel File and select GenericAssay_Run1.xls. (Wait a few seconds for the file to upload.)
- Notice that the Created and Modified fields are filled in automatically, as specified in the module-based assay's upload.html file.
- Click Import Data and repeat the import process for GenericAssay_Run2.xls.
- Click Done.
Review Imported Data
- Click on the first run (GenericAssayRun1.xls) to see the data it contains. You will see data similar to the following:
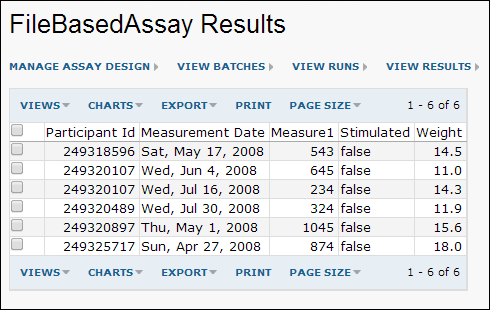
- You can now integrate this data into any available target studies.
Related Topics
Assay Custom Domains
An assay module can define a custom domain to replace LabKey's built-in default assay domains, by adding a schema definition in the domains/ directory. For example:
assay/<assay-name>/domains/<domain-name>.xml
The name of the assay is taken from the <assay-name> directory. The contents of <domain-name>.xml file contains the domain definition and conforms to the <domain> element from assayProvider.xsd, which is in turn a DomainDescriptorType from the expTypes.xsd XML schema. There are three built-in domains for assays: "batch", "run", and "result". This following result domain replaces the build-in result domain for assays:
result.xml
<ap:domain xmlns:exp="http://cpas.fhcrc.org/exp/xml"
xmlns:ap="http://labkey.org/study/assay/xml">
<exp:Description>This is my data domain.</exp:Description>
<exp:PropertyDescriptor>
<exp:Name>SampleId</exp:Name>
<exp:Description>The Sample Id</exp:Description>
<exp:Required>true</exp:Required>
<exp:RangeURI>http://www.w3.org/2001/XMLSchema#string</exp:RangeURI>
<exp:Label>Sample Id</exp:Label>
</exp:PropertyDescriptor>
<exp:PropertyDescriptor>
<exp:Name>TimePoint</exp:Name>
<exp:Required>true</exp:Required>
<exp:RangeURI>http://www.w3.org/2001/XMLSchema#dateTime</exp:RangeURI>
</exp:PropertyDescriptor>
<exp:PropertyDescriptor>
<exp:Name>DoubleData</exp:Name>
<exp:RangeURI>http://www.w3.org/2001/XMLSchema#double</exp:RangeURI>
</exp:PropertyDescriptor>
</ap:domain>
To deploy the module, the assay directory is zipped up as a <module-name>.module file and copied to the LabKey server's modules directory.
When you create a new assay design for that assay type, it will use the fields defined in the XML domain as a template for the corresponding domain. Changes to the domains in the XML files will not affect existing assay designs that have already been created.
Assay Custom Views
Add a Custom Details View
Suppose you want to add a [details] link to each row of an assay run table, that takes you to a custom details view for that row. You can add new views to the module-based assay by adding html files in the views/ directory, for example:
assay/<assay-name>/views/<view-name>.html
The overall page template will include JavaScript objects as context so that they're available within the view, avoiding an extra client API request to fetch it from the server. For example, the result.html page can access the assay definition and result data as LABKEY.page.assay and LABKEY.page.result respectively. Here is an example custom details view named result.html:
1 <table>
2 <tr>
3 <td class='labkey-form-label'>Sample Id</td>
4 <td><div id='SampleId_div'>???</div></td>
5 </tr>
6 <tr>
7 <td class='labkey-form-label'>Time Point</td>
8 <td><div id='TimePoint_div'>???</div></td>
9 </tr>
10 <tr>
11 <td class='labkey-form-label'>Double Data</td>
12 <td><div id='DoubleData_div'>???</div></td>
13 </tr>
14 </table>
15
16 <script type="text/javascript">
17 function setValue(row, property)
18 {
19 var div = Ext.get(property + "_div");
20 var value = row[property];
21 if (!value)
22 value = "<none>";
23 div.dom.innerHTML = value;
24 }
25
26 if (LABKEY.page.result)
27 {
28 var row = LABKEY.page.result;
29 setValue(row, "SampleId");
30 setValue(row, "TimePoint");
31 setValue(row, "DoubleData");
32 }
33 </script>
Note on line 28 the details view is accessing the result data from LABKEY.page.result. See Example Assay JavaScript Objects for a description of the LABKEY.page.assay and LABKEY.page.result objects.
Add a custom view for a run
Same as for the custom details page for the row data except the view file name is run.html and the run data will be available as the LABKEY.page.run variable. See Example Assay JavaScript Objects for a description of the LABKEY.page.run object.
Add a custom view for a batch
Same as for the custom details page for the row data except the view file name is batch.html and the run data will be available as the LABKEY.page.batch variable. See Example Assay JavaScript Objects for a description of the LABKEY.page.batch object.
Example Assay JavaScript Objects
LABKEY.page.assay:
The assay definition is available as LABKEY.page.assay for all of the html views. It is a JavaScript object, which is of type LABKEY.Assay.AssayDesign:
LABKEY.page.assay = {
"id": 4,
"projectLevel": true,
"description": null,
"name": <assay name>,
// domains objects: one for batch, run, and result.
"domains": {
// array of domain property objects for the batch domain
"<assay name> Batch Fields": [
{
"typeName": "String",
"formatString": null,
"description": null,
"name": "ParticipantVisitResolver",
"label": "Participant Visit Resolver",
"required": true,
"typeURI": "http://www.w3.org/2001/XMLSchema#string"
},
{
"typeName": "String",
"formatString": null,
"lookupQuery": "Study",
"lookupContainer": null,
"description": null,
"name": "TargetStudy",
"label": "Target Study",
"required": false,
"lookupSchema": "study",
"typeURI": "http://www.w3.org/2001/XMLSchema#string"
}
],
// array of domain property objects for the run domain
"<assay name> Run Fields": [{
"typeName": "Double",
"formatString": null,
"description": null,
"name": "DoubleRun",
"label": null,
"required": false,
"typeURI": "http://www.w3.org/2001/XMLSchema#double"
}],
// array of domain property objects for the result domain
"<assay name> Result Fields": [
{
"typeName": "String",
"formatString": null,
"description": "The Sample Id",
"name": "SampleId",
"label": "Sample Id",
"required": true,
"typeURI": "http://www.w3.org/2001/XMLSchema#string"
},
{
"typeName": "DateTime",
"formatString": null,
"description": null,
"name": "TimePoint",
"label": null,
"required": true,
"typeURI": "http://www.w3.org/2001/XMLSchema#dateTime"
},
{
"typeName": "Double",
"formatString": null,
"description": null,
"name": "DoubleData",
"label": null,
"required": false,
"typeURI": "http://www.w3.org/2001/XMLSchema#double"
}
]
},
"type": "Simple"
};LABKEY.page.batch:
The batch object is available as LABKEY.page.batch on the upload.html and batch.html pages. The JavaScript object is an instance of LABKEY.Exp.RunGroup and is shaped like:
LABKEY.page.batch = new LABKEY.Exp.RunGroup({
"id": 8,
"createdBy": <user name>,
"created": "8 Apr 2009 12:53:46 -0700",
"modifiedBy": <user name>,
"name": <name of the batch object>,
"runs": [
// array of LABKEY.Exp.Run objects in the batch. See next section.
],
// map of batch properties
"properties": {
"ParticipantVisitResolver": null,
"TargetStudy": null
},
"comment": null,
"modified": "8 Apr 2009 12:53:46 -0700",
"lsid": "urn:lsid:labkey.com:Experiment.Folder-5:2009-04-08+batch+2"
});
LABKEY.page.run:
The run detail object is available as LABKEY.page.run on the run.html pages. The JavaScript object is an instance of LABKEY.Exp.Run and is shaped like:
LABKEY.page.run = new LABKEY.Exp.Run({
"id": 4,
// array of LABKEY.Exp.Data objects added to the run
"dataInputs": [{
"id": 4,
"created": "8 Apr 2009 12:53:46 -0700",
"name": "run01.tsv",
"dataFileURL": "file:/C:/Temp/assaydata/run01.tsv",
"modified": null,
"lsid": <filled in by the server>
}],
// array of objects, one for each row in the result domain
"dataRows": [
{
"DoubleData": 3.2,
"SampleId": "Monkey 1",
"TimePoint": "1 Nov 2008 11:22:33 -0700"
},
{
"DoubleData": 2.2,
"SampleId": "Monkey 2",
"TimePoint": "1 Nov 2008 14:00:01 -0700"
},
{
"DoubleData": 1.2,
"SampleId": "Monkey 3",
"TimePoint": "1 Nov 2008 14:00:01 -0700"
},
{
"DoubleData": 1.2,
"SampleId": "Monkey 4",
"TimePoint": "1 Nov 2008 00:00:00 -0700"
}
],
"createdBy": <user name>,
"created": "8 Apr 2009 12:53:47 -0700",
"modifiedBy": <user name>,
"name": <name of the run>,
// map of run properties
"properties": {"DoubleRun": null},
"comment": null,
"modified": "8 Apr 2009 12:53:47 -0700",
"lsid": "urn:lsid:labkey.com:SimpleRun.Folder-5:cf1fea1d-06a3-102c-8680-2dc22b3b435f"
});
LABKEY.page.result:
The result detail object is available as LABKEY.page.result on the result.html page. The JavaScript object is a map for a single row and is shaped like:
LABKEY.page.result = {
"DoubleData": 3.2,
"SampleId": "Monkey 1",
"TimePoint": "1 Nov 2008 11:22:33 -0700"
};Assay Query Metadata
Query Metadata for Assay Tables
You can associate query metadata with an individual assay design, or all assay designs that are based on the same type of assay (e.g., "NAb" or "Viability").
Example. Assay table names are based upon the name of the assay design. For example, consider an assay design named "Example" that is based on the "Viability" assay type. This design would be associated with three tables in the schema explorer: "Example Batches", "Example Runs", and "Example Data."
Associate metadata with a single assay design. To attach query metadata to the "Example Data" table, you would normally create a /queries/assay/Example Data.query.xml metadata file. This would work well for the "Example Data" table itself. However, this method would not allow you to re-use this metadata file for a new assay design that is also based on the same assay type ("Viability" in this case).
Associate metadata with all assay designs based on a particular assay type. To permit re-use of the metadata, you need to create a query metadata file whose name is based upon the assay type and table name. To continue our example, you would create a query metadata file callled /assay/Viability/queries/Data.query.xml to attach query metadata to all data tables based on the Viability-type assay.
As with other query metadata in module files, the module must be activated (in other words, the appropriate checkbox must be checked) in the folder's settings.
See Modules: Queries, Views and Reports and Modules: Query Metadata for more information on query metadata.
Customize Batch Save Behavior
The AssaySaveHandler interface enables file-based assays to extend the functionality of the SaveAssayBatch action with Java code. A file-based assay can provide an implementation of this interface by creating a Java-based module and then putting the class under the module's src directory. This class can then be referenced by name in the <saveHandler/> element in the assay's config file. For example, an entry might look like:
<saveHandler>org.labkey.icemr.assay.tracking.TrackingSaveHandler</saveHandler>.
To implement this functionality:
- Create the skeleton framework for a Java module. This consists of a controller class, manager, etc. See Creating a New Java Module for details on autogenerating the boiler plate Java code.
- Add an assay directory underneath the Java src directory that corresponds to the file-based assay you want to extend. For example: myModule/src/org.labkey.mymodule/assay/tracking
- Implement the AssaySaveHandler interface. You can choose to either implement the interface from scratch or extend default behavior by having your class inherit from the DefaultAssaySaveHandler class. If you want complete control over the JSON format of the experiment data you want to save, you may choose to implement the AssaySaveHandler interface entirely. If you want to follow the pre-defined LABKEY experiment JSON format, then you can inherit from the DefaultAssaySaveHandler class and only override the specific piece you want to customize. For example, you may want custom code to run when a specific property is saved. (See below for more implementation details.)
- Reference your class in the assay's config.xml file. For example, notice the <ap:saveHandler/> entry below. If a non-fully-qualified name is used (as below) then LabKey Server will attempt to find this class under org.labkey.[module name].assay.[assay name].[save handler name].
<ap:provider xmlns:ap="http://labkey.org/study/assay/xml">
<ap:name>Flask Tracking</ap:name>
<ap:description>
Enables entry of a set of initial samples and then tracks
their progress over time via a series of daily measurements.
</ap:description>
<ap:saveHandler>TrackingSaveHandler</ap:saveHandler>
<ap:fieldKeys>
<ap:participantId>Run/PatientId</ap:participantId>
<ap:date>MeasurementDate</ap:date>
</ap:fieldKeys>
</ap:provider>
- The interface methods are invoked when the user chooses to import data into the assay or otherwise calls the SaveAssayBatch action. This is usually invoked by the Experiment.saveBatch JavaScript API. On the server, the file-based assay provider will look for an AssaySaveHandler specified in the config.xml and invoke its functions. If no AssaySaveHandler is specfied then the DefaultAssaySaveHandler implementation is used.
SaveAssayBatch Details
The SaveAssayBatch function creates a new instance of the SaveHandler for each request. SaveAssayBatch will dispatch to the methods of this interface according to the format of the JSON Experiment Batch (or run group) sent to it by the client. If a client chooses to implement this interface directly then the order of method calls will be:
- beforeSave
- handleBatch
- afterSave
- beforeSave
- handleBatch
- handleProperties (for the batch)
- handleRun (for each run)
- handleProperties (for the run)
- handleProtocolApplications
- handleData (for each data output)
- handleProperties (for the data)
- handleMaterial (for each input material)
- handleProperties (for the material)
- handleMaterial (for each output material)
- handleProperties (for the material)
- afterSave
Related Topics
SQL Scripts for Module-Based Assays
Some options:
1) Manually import a list archive into the target folder.
2) Add the tables via SQL scripts included in the module. To insert data: use SQL DML scripts or create an initialize.html view that populates the table using LABKEY.Query.insertRows().
To add the supporting table using SQL scripts, add a schemas directory, as a sibling to the assay directory, as shown below.
exampleassay
├───assay
│ └───example
│ │ config.xml
│ │
│ ├───domains
│ │ batch.xml
│ │ result.xml
│ │ run.xml
│ │
│ └───views
│ upload.html
│
└───schemas
│ SCHEMA_NAME.xml
│
└───dbscripts
├───postgresql
│ SCHEMA_NAME-X.XX-Y.YY.sql
└───sqlserver
SCHEMA_NAME-X.XX-Y.YY.sql
To support only one database, include a script only for that database, and configure your module properties accordingly -- see "SupportedDatabases" in Module Properties Reference.
LabKey Server does not currently support adding assay types or lists via SQL scripts, but you can create a new schema to hold the table, for example, the following script creates a new schema called "myreagents" (on PostgreSQL):
DROP SCHEMA IF EXISTS myreagents CASCADE;
CREATE SCHEMA myreagents;
CREATE TABLE myreagents.Reagents
(
RowId SERIAL NOT NULL,
ReagentName VARCHAR(30) NOT NULL
);
ALTER TABLE ONLY myreagents.Reagents
ADD CONSTRAINT Reagents_pkey PRIMARY KEY (RowId);
INSERT INTO myreagents.Reagents (ReagentName) VALUES ('Acetic Acid');
INSERT INTO myreagents.Reagents (ReagentName) VALUES ('Baeyers Reagent');
INSERT INTO myreagents.Reagents (ReagentName) VALUES ('Carbon Disulfide');
Update the assay domain, adding a lookup/foreign key property to the Reagents table:
<exp:PropertyDescriptor>
<exp:Name>Reagent</exp:Name>
<exp:Required>false</exp:Required>
<exp:RangeURI>http://www.w3.org/2001/XMLSchema#int</exp:RangeURI>
<exp:Label>Reagent</exp:Label>
<exp:FK>
<exp:Schema>myreagents</exp:Schema>
<exp:Query>Reagents</exp:Query>
</exp:FK>
</exp:PropertyDescriptor>
If you'd like to allow admins to add/remove fields from the table, you can add an LSID column to your table and make it a foreign key to the exp.Object.ObjectUri column in the schema.xml file. This will allow you to define a domain for the table much like a list. The domain is per-folder so different containers may have different sets of fields.
For example, see customModules/reagent/resources/schemas/reagent.xml. It wires up the LSID lookup to the exp.Object.ObjectUri column
<ns:column columnName="Lsid">
<ns:datatype>lsidtype</ns:datatype>
<ns:isReadOnly>true</ns:isReadOnly>
<ns:isHidden>true</ns:isHidden>
<ns:isUserEditable>false</ns:isUserEditable>
<ns:isUnselectable>true</ns:isUnselectable>
<ns:fk>
<ns:fkColumnName>ObjectUri</ns:fkColumnName>
<ns:fkTable>Object</ns:fkTable>
<ns:fkDbSchema>exp</ns:fkDbSchema>
</ns:fk>
</ns:column>
...and adds an "Edit Fields" button that opens the domain editor.
function editDomain(queryName)
{
var url = LABKEY.ActionURL.buildURL("property", "editDomain", null, {
domainKind: "ExtensibleTable",
createOrEdit: true,
schemaName: "myreagents",
queryName: queryName
});
window.location = url;
}
Transformation Scripts
Topics
- Use Transformation Scripts
- How Transformation Scripts Work
- Example Workflow: Develop a Transformation Script (perl)
- Example Transformation Scripts (perl)
- Transformation Scripts in R
- Transformation Scripts in Java
- Transformation Scripts for Module-based Assays
- Run Properties Reference
- Transformation Script Substitution Syntax
- Warnings in Tranformation Scripts
Use Transformation Scripts
Each assay design can be associated with one or more validation or transformation scripts which are run in the order specified. The script file extension (.r, .pl, etc) identifies the script engine that will be used to run the transform script. For example: a script named test.pl will be run with the Perl scripting engine. Before you can run validation or transformation scripts, you must configure the necessary Scripting Engines.
To specify a transform script in an assay design, you enter the full path including the file extension.
- Open the assay designer for a new assay, or edit an existing assay design.
- Click Add Script.
- Enter the full path to the script in the Transform Scripts field.
- You may enter multiple scripts by clicking Add Script again.
- Confirm that other Properties required by your assay type are correctly specified.
- Click Save and Close.
When you import (or re-import) run data using this assay design, the script will be executed. When you are developing or debugging transform scripts, you can use the Save Script Data option to store the files generated by the server that are passed to the script. Once your script is working properly, uncheck this box to avoid unnecessarily cluttering your disk.
A few notes on usage:
- Client API calls are not supported in transform scripts.
- Columns populated by transform scripts must already exist in the assay definition.
- Executed scripts show up in the experimental graph, providing a record that transformations and/or quality control scripts were run.
- Transform scripts are run before field-level validators.
- The script is invoked once per run upload.
- Multiple scripts are invoked in the order they are listed in the assay design.
The general purpose assay tutorial includes another example use of a transformation script in Set up a Data Transformation Script.
How Transformation Scripts Work
Script Execution Sequence
Transformation and validation scripts are invoked in the following sequence:
- A user uploads assay data.
- The server creates a runProperties.tsv file and rewrites the uploaded data in TSV format. Assay-specific properties and files from both the run and batch levels are added. See Run Properties Reference for full lists of properties.
- The server invokes the transform script by passing it the information created in step 2 (the runProperties.tsv file).
- After script completion, the server checks whether any errors have been written by the transform script and whether any data has been transformed.
- If transformed data is available, the server uses it for subsequent steps; otherwise, the original data is used.
- If multiple transform scripts are specified, the server invokes the other scripts in the order in which they are defined.
- Field-level validator/quality-control checks (including range and regular expression validation) are performed. (These field-level checks are defined in the assay definition.)
- If no errors have occurred, the run is loaded into the database.
Passing Run Properties to Transformation Scripts
Information on run properties can be passed to a transform script in two ways. You can put a substitution token into your script to identify the run properties file, or you can configure your scripting engine to pass the file path as a command line argument. See Transformation Script Substitution Syntax for a list of available substitution tokens.
For example, using perl:
Option #1: Put a substitution token (${runInfo}) into your script and the server will replace it with the path to the run properties file. Here's a snippet of a perl script that uses this method:
# Open the run properties file. Run or upload set properties are not used by
# this script. We are only interested in the file paths for the run data and
# the error file.
open my $reportProps, '${runInfo}';
Option #2: Configure your scripting engine definition so that the file path is passed as a command line argument:
- Go to Admin > Site > Admin Console.
- Select the Views and Scripting.
- Select and edit the perl engine.
- Add ${runInfo} to the Program Command field.
Example Workflow: Develop a Transformation Script (perl)
- transform run data
- transform run properties
Script Engine Setup
Before you can develop or run validation or transform scripts, configure the necessary Scripting Engines. You only need to set up a scripting engine once per type of script. You will need a copy of Perl running on your machine to set up the engine.
- Select Admin > Site > Admin Console.
- Click Views and Scripting.
- Click Add > New Perl Engine.
- Fill in as shown, specifying the "pl" extension and full path to the perl executable.
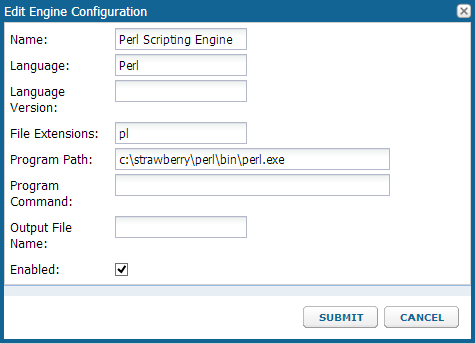
- Click Submit.
Add a Script to the Assay Design
Create a new empty .pl file in the development location of your choice and include it in your assay design.
- Navigate to the Assay Tutorial.
- Click GenericAssay in the Assay List web part.
- Select Manage Assay Design > copy assay design.
- Click Copy to Current Folder.
- Enter a new name, such as "TransformedAssay".
- Click Add Script and type the full path to the new script file you are creating.
- Check the box for Save Script Data.
- Confirm that the batch, run, and data fields are correct.
- Click Save and Close.
Obtain Test Data
To assist in writing your transform script, you will next obtain sample "runData.tsv" and "runProperties.tsv" files showing the state of your data import 'before' the transform script would be applied. To generate useful test data, you need to import a data run using the new assay design.
- Open and select the following file (if you have already imported this file during the tutorial, you will first need to delete that run):
LabKeyDemoFiles/Assays/Generic/GenericAssay_Run4.xls
- Click Import Data.
- Select the TransformedAssay design you just defined, then click Import.
- Click Next, then Save and Finish.
- When the import completes, select Manage Assay Design > edit assay design.
- You will now see a Download Test Data button that was not present during initial assay design.
- Click it and unzip the downloaded "sampleQCData" package to see the .tsv files.
- Open the "runData.tsv" file to view the current fields.
Date VisitID ParticipantID M3 M2 M1 SpecimenID
12/17/2013 1234 demo value 1234 1234 1234 demo value
12/17/2013 1234 demo value 1234 1234 1234 demo value
12/17/2013 1234 demo value 1234 1234 1234 demo value
12/17/2013 1234 demo value 1234 1234 1234 demo value
12/17/2013 1234 demo value 1234 1234 1234 demo value
Save Script Data
Typically transform and validation script data files are deleted on script completion. For debug purposes, it can be helpful to be able to view the files generated by the server that are passed to the script. When the Save Script Data checkbox is checked, files will be saved to a subfolder named: "TransformAndValidationFiles", in the same folder as the original script. Beneath that folder are subfolders for the AssayId, and below that a numbered directory for each run. In that nested subdirectory you will find a new "runDataFile.tsv" that will contain values from the run file plugged into the current fields.
participantid Date M1 M2 M3
249318596 2008-06-07 00:00 435 1111 15.0
249320107 2008-06-06 00:00 456 2222 13.0
249320107 2008-03-16 00:00 342 3333 15.0
249320489 2008-06-30 00:00 222 4444 14.0
249320897 2008-05-04 00:00 543 5555 32.0
249325717 2008-05-27 00:00 676 6666 12.0
Define the Desired Transformation
The runData.tsv file gives you the basic fields layout. Decide how you need to modify the default data. For example, perhaps for our project we need an adjusted version of the value in the M1 field - we want the doubled value available as an integer.
Add Required Fields to the Assay Design
- Select Manage Assay Design > edit assay design.
- Scroll down to the Data Fields section and click Add Field.
- Enter "AdjustM1", "Adjusted M1", and select type "Integer".
- Click Save and Close.
Write a Script to Transform Run Data
Now you have the information you need to write and refine your transformation script. Open the empty script file and paste the contents of the Modify Run Data box from this page: Example Transformation Scripts (perl).
Iterate over the Sample Run
Re-import the same run using the transform script you have defined.
- From the run list, select the run and click Re-import Run.
- Click Next.
- Under Run Data, click Use the data file(s) already uploaded to the server.
- Click Save and Finish.
The results now show the new field populated with the Adjusted M1 value.
Until the results are as desired, you will edit the script and use Reimport Run to retry.
Once your transformation script is working properly, re-edit the assay design one more time to uncheck the Save Script Data box - otherwise your script will continue to generate artifacts with every run and could eventually fill your disk.
Debugging Transformation Scripts
If your script has errors that prevent import of the run, you will see red text in the Run Properties window. If you fail to select the correct data file, for example:
If you have a type mismatch error between your script results and the defined destination field, you will see an error like:
Errors File
If the validation script needs to report an error that is displayed by the server, it adds error records to an error file. The location of the error file is specified as a property entry in the run properties file. The error file is in a tab-delimited format with three columns:
- type: error, warning, info, etc.
- property: (optional) the name of the property that the error occurred on.
- message: the text message that is displayed by the server.
| type | property | message |
|---|---|---|
| error | runDataFile | A duplicate PTID was found : 669345900 |
| error | assayId | The assay ID is in an invalid format |
Example Transformation Scripts (perl)
- Modify Run Data
- Modify Run Properties
Modify Run Data
This script is used in the Example Workflow: Develop a Transformation Script (perl) and populates a new field with data derived from an existing field in the run.
#!/usr/local/bin/perl
use strict;
use warnings;
# Open the run properties file. Run or upload set properties are not used by
# this script. We are only interested in the file paths for the run data and
# the error file.
open my $reportProps, '${runInfo}';
my $transformFileName = "unknown";
my $dataFileName = "unknown";
my %transformFiles;
# Parse the data file properties from reportProps and save the transformed data location
# in a map. It's possible for an assay to have more than one transform data file, although
# most will only have a single one.
while (my $line=<$reportProps>)
{
chomp($line);
my @row = split(/t/, $line);
if ($row[0] eq 'runDataFile')
{
$dataFileName = $row[1];
# transformed data location is stored in column 4
$transformFiles= $row[3];
}
}
my $key;
my $value;
my $adjustM1 = 0;
# Read each line from the uploaded data file and insert new data (double the value in the M1 field)
# into an additional column named 'Adjusted M1'. The additional column must already exist in the assay
# definition and be of the correct type.
while (($key, $value) = each(%transformFiles)) {
open my $dataFile, $key or die "Can't open '$key': $!";
open my $transformFile, '>', $value or die "Can't open '$value': $!";
my $line=<$dataFile>;
chomp($line);
$line =~ s/r*//g;
print $transformFile $line, "\t", "Adjusted M1", "\n";
while (my $line=<$dataFile>)
{
$adjustM1 = substr($line, 27, 3) * 2;
chomp($line);
$line =~ s/r*//g;
print $transformFile $line, "\t", $adjustM1, "\n";
}
close $dataFile;
close $transformFile;
}
Modify Run Properties
You can also define a transform script that modifies the run properties, as show in this example which parses the short filename out of the full path:
#!/usr/local/bin/perl
use strict;
use warnings;
# open the run properties file, run or upload set properties are not used by
# this script, we are only interested in the file paths for the run data and
# the error file.
open my $reportProps, $ARGV[0];
my $transformFileName = "unknown";
my $uploadedFile = "unknown";
while (my $line=<$reportProps>)
{
chomp($line);
my @row = split(/\t/, $line);
if ($row[0] eq 'transformedRunPropertiesFile')
{
$transformFileName = $row[1];
}
if ($row[0] eq 'runDataUploadedFile')
{
$uploadedFile = $row[1];
}
}
if ($transformFileName eq 'unknown')
{
die "Unable to find the transformed run properties data file";
}
open my $transformFile, '>', $transformFileName or die "Can't open '$transformFileName': $!";
#parse out just the filename portion
my $i = rindex($uploadedFile, "\\") + 1;
my $j = index($uploadedFile, ".xls");
#add a value for fileID
print $transformFile "FileID", "\t", substr($uploadedFile, $i, $j-$i), "\n";
close $transformFile;
Transformation Scripts in R
Overview
Users importing instrument-generated tabular datasets into LabKey Server may run into the following difficulties:
- Instrument-generated files often contain header lines before the main dataset, denoted by a leading # or ! or other symbol. These lines usually contain useful metadata about the protocol or reagents or samples tested, and in any case need to be skipped over to find the main data set.
- The file format is optimized for display, not for efficient storage and retrieval. For example, columns that correspond to individual samples are difficult to work with in a database.
- The data to be imported contains the display values from a lookup column, which need to be mapped to the foreign key values for storage.
First we review the way to hookup a transform script to an assay and the communications mechanisms between the assay framework and a transform script in R.
Identifying the Path to the Script File
Transform scripts are designated as part of a assay by providing a fully qualified path to the script file in the field named at the top of the assay instance definition. A convenient location to put the script file is to upload it using a File web part defined in the same folder as the assay definition. Then the fully qualified path to the script file is the concatenation of the file root for the folder (for example, "C:\lktrunk\build\deploy\files\MyAssayFolderName\@files\", as determined by the Files page in the Admin console) plus the file path to the script file as seen in the File web part (for example, "scripts\LoadData.R". For the file path, LabKey Server accepts the use of either backslashes (the default Windows format) or forward slashes.
When working on your own developer workstation, you can put the script file wherever you like, but putting it within the scope of the File manager will make it easier to deploy to a server. It also makes iterative development against a remote server easier, since you can use a Web-DAV enabled file editor to directly edit the same file that the server is calling.
If your transform script calls other script files to do its work, the normal way to pull in the source code is using the source statement, for example
source("C:\lktrunk\build\deploy\files\MyAssayFolderName\@files\Utils.R")But to keep the scripts so that they are easily moved to other servers, it is better to keep the script files together and the built-in substitution token "${srcDirectory}" which the server automatically fills in to be the directory where the called script file is located , for example:
source("${srcDirectory}/Utils.R");Accessing and Using the Run Properties File
The primary mechanism for communication between the LabKey Assay framework and the Transform script is the Run Properties file. Again a substitution token tells the script code where to find this file. The script file should contain a line like
rpPath<- "${runInfo}"When the script is invoked by the assay framework, the rpPath variable will contain the fully qualified path to the run properties file.
The run properties file contains three categories of properties:
1. Batch and run properties as defined by the user when creating an assay instance. These properties are of the format: <property name> <property value> <java data type>
for example,
gDarkStdDev 1.98223 java.lang.DoubleWhen the transform script is called these properties will contain any values that the user has typed into the corresponding text box under the “Batch Properties” of “Run Properties” sections of the upload form. The transform script can assign or modify these properties based on calculations or by reading them from the raw data file from the instrument. The script must then write the modified properties file to the location specified by the transformedRunPropertiesFile property (see #3 below)
2. Context properties of the assay such as assayName, runComments, and containerPath. These are recorded in the same format as the user-defined batch and run properties, but they cannot be overwritten by the script.
3. Paths to input and output files. These are fully qualified paths that the script reads from or writes to. They are in a <property name> <property value> format without property types. The paths currently used are:
- a. runDataUploadedFile: the raw data file that was selected by the user and uploaded to the server as part of an import process. This can be an Excel file, a tab-separated text file, or a comma-separated text file.
- b. runDataFile: the imported data file after the assay framework has attempted to convert the file to .tsv format and match its columns to the assay data result set definition. The path will point to a subfolder below the script file directory, with a path value similar to <property value> <java property type>. The AssayId_22\42 part of the directory path serves to separate the temporary files from multiple executions by multiple scripts in the same folder.
C:\lktrunk\build\deploy\files\transforms\@files\scripts\TransformAndValidationFiles\AssayId_22\42\runDataFile.tsv
- c. AssayRunTSVData: This file path is where the result of the transform script will be written. It will point to a unique file name in an “assaydata” directory that the framework creates at the root of the files tree. NOTE: this property is written on the same line as the runDataFile property.
- d. errorsFile: This path is where a transform or validation script can write out error messages for use in troubleshooting. Not normally needed by an R script because the script usually writes errors to stdout, which are written by the framework to a file named “<scriptname>.Rout”.
- e. transformedRunPropertiesFile: This path is where the script writes out the updated values of batch- and run-level properties that are listed in the runProperties file.
Choosing the Input File for Transform Script Processing
The transform script developer can choose to use either the runDataFile or the runDataUploadedFile as its input. The runDataFile would be the right choice for an Excel-format raw file and a script that fills in additional columns of the data set. By using the runDataFile, the assay framework does the Excel-to-TSV conversion and the script doesn’t need to know how to parse Excel files. The runDataUploadedFile would be the right choice for a raw file in TSV format that the script is going to reformat by turning columns into rows. In either case, the script writes its output to the AssayRunTSVData file.
Transform Script Options
There are two useful options presented as checkboxes in the Assay designer.
- Save Script Data tells the framework to not delete the intermediate files such as the runProperties file after a successful run. This option is important during script development. It can be turned off to avoid cluttering the file space under the TransformAndValidationFiles directory that the framework automatically creates under the script file directory.
- Upload In Background tells the framework to create a pipeline job as part of the import process, rather than tying up the browser session. It is useful for importing large data sets.
Connecting Back to the Server from a Transform Script
Sometimes a transform script needs to connect back to the server to do its job. One example is translating lookup display values into key values. The Rlabkey library available on CRAN has the functions needed to connect to, query, and insert or update data in the local LabKey Server where it is running. To give the connection the right security context (the current user’s), the assay framework provides the substitution token ${rLabkeySessionId}. Including this token on a line by itself near the beginning of the transform script eliminates the need to use a config file to hold a username and password for this loopback connection. It will be replaced with two lines that looks like:
labkey.sessionCookieName = "JSESSIONID" labkey.sessionCookieContents = "TOMCAT_SESSION_ID"
where TOMCAT_SESSION_ID is the actual ID of the user's HTTP session.
Debugging an R Transform Script
You can load an R transform script into the R console/debugger and run the script with debug(<functionname> commands active. Since the substitution tokens described above ( ${srcDirectory} , ${runInfo}, and ${rLabkeySessionId} ) are necessary to the correct operation of the script, the framework conveniently writes out a version of the script with these substitutions made, into the same subdiretory as the runProperties.tsv file is found. Load this modified version of the script into the R console.
Example Script
Setup
- Create a new project, type Assay
- Add the following Web parts:
- Files
- Lists
- Data Pipeline
- Sample Sets (narrow)
- Copy the scripts folder from the data folder to the root of the Files web part tree
- Create a sample set called ExampleSamples
- Click on header of Sample Sets web part
- Select Import Sample Set
- Open the file samples.txt in a text editor or Excel (Click to download from page.)
- Copy and paste the contents into the import window , select sampleId as the key field
- Create a list called probesources by importing ProbeSourcesListArchive.zip (Click to download.)
- Create a GPAT assay with transform script
- Import the file ExampleAssayWithRTransform.xar (Click to download.)
- Fix up the path to the script file TransformScriptExample.R (Click to download.)
- Run the assay
- Click on assay name
- Import data button on toolbar
- Select probe source from list, leave property Prefix, press Next
- Column Names Row: 65
- Sample Set: ExampleSamples
- Run Data: Upload a data file. Choose file GSE11199_series_matrix_200.txt (Click to download.)
- Save and finish
A Look at the Code
This transform script example handles the data output from an Affymetrics microarray reader. The data file contains 64 lines of metadata before the chip-level intensity data. The metadata describes the platform, the experiment, and the samples used. The spot-level data is organized with one column per sample, which may be efficient for storage in a spreadsheet but isn’t good for querying in a database.
The transform script does the following tasks:
- Reads in the runProperties file
- Gets additional import processing parameters from a lookup list, such as the prefix that designates a comment line containing a property-value pair
- Fills in run properties that are read from the data file header (marked by the prefix). Writes the transformed run properties to the designated file location so they get stored with the assay.
- Converts sample identifiers to sample set key values so that a lookup from result data to sample set properties works.
- Skips over a specified number of rows to the beginning of the spot data.
- Reshapes the input data so that the result set is easier to query by sample
TransformScriptExample.R
options(stringsAsFactors = FALSE)
source("${srcDirectory}/ExampleUtils.R")
baseUrl<-"http://localhost:8080/labkey"
${rLabkeySessionId}
rpPath<- "${runInfo}"
## read the file paths etc out of the runProperties.tsv file
params <- getRunPropsList(rpPath, baseUrl)
## read the input data frame just to get the column headers.
inputDF<-read.table(file=params$inputPathUploadedFile, header = TRUE,
sep = "\t", quote = "\"",
fill=TRUE, stringsAsFactors = FALSE, check.names=FALSE,
row.names=NULL, skip=(params$loaderColNamesRow -1), nrows=1)
cols<-colnames(inputDF)
## create a Name to RowId map for samples
keywords <- as.vector(colnames(inputDF)[-1])
queryName=params$sampleSetName
keywordMap<- getLookupMap( keywords, baseUrl=baseUrl, folderPath=params$containerPath,
schemaName="Samples", queryName=queryName, keyField="rowId",
displayField="SampleId")
doRunLoad(params=params, inputColNames=cols, outputColNames=c( "ID_REF", "sample", "val"),
lookupMap=keywordMap)
ExampleUtils.R, function getRunPropsList()
getRunPropsList<- function(rpPath, baseUrl)
{
rpIn<- read.table(rpPath, col.names=c("name", "val1", "val2", "val3"), #########
header=FALSE, check.names=FALSE, ## 1 ##
stringsAsFactors=FALSE, sep="\t", quote="", fill=TRUE, na.strings=""); #########
## pull out the run properties
params<- list(inputPathUploadedFile = rpIn$val1[rpIn$name=="runDataUploadedFile"],
inputPathValidated = rpIn$val1[rpIn$name=="runDataFile"],
##a little strange. AssayRunTSVData is the one we need to output to
outputPath = rpIn$val3[rpIn$name=="runDataFile"],
containerPath = rpIn$val1[rpIn$name=="containerPath"],
runPropsOutputPath = rpIn$val1[rpIn$name=="transformedRunPropertiesFile"],
sampleSetId = as.integer(rpIn$val1[rpIn$name=="sampleSet"]),
probeSourceId = as.integer(rpIn$val1[rpIn$name=="probeSource"]),
errorsFile = rpIn$val1[rpIn$name=="errorsFile"])
## lookup the name of the sample set based on its number
if (length(params$sampleSetId)>0)
{
df<-labkey.selectRows(baseUrl=baseUrl,
folderPath=params$containerPath, schemaName="exp", queryName="SampleSets",
colFilter=makeFilter(c("rowid", "EQUALS", params$sampleSetId)))
params<- c(params, list(sampleSetName=df$Name))
}
## This script reformats the rows in batches of 1000 in order to reduce
## the memory requirements of the R calculations
params<-c(params, list(loaderBatchSize=as.integer(1000)))
## From the probesource lookup table, get the prefix characters that
## identify property value comment lines in the data file, and the starting
## line number of the spot data table within the data file
dfProbeSource=labkey.selectRows(baseUrl=baseUrl, folderPath=params$containerPath, #########
schemaName="lists", queryName="probesources", ## 2 ##
colFilter=makeFilter(c("probesourceid", "EQUALS", params$probeSourceId))) #########
params<-c(params, list(propertyPrefix=dfProbeSource$propertyPrefix,
loaderColNamesRow=dfProbeSource$loaderColNamesRow))
if (is.null(params$loaderColNamesRow) | is.na(params$loaderColNamesRow))
{
params$loaderColNamesRow <- 1
}
## now apply the run property values reported in the header
## of the data tsv file to the corresponding run properties
conInput = file(params$inputPathUploadedFile, "r")
line<-""
pfx <- as.integer(0)
fHasProps <- as.logical(FALSE)
if (!is.na(params$propertyPrefix))
{ #########
pfx<-nchar(params$propertyPrefix) ## 3 ##
} #########
while(pfx>0)
{
line<-readLines(conInput, 1)
if (nchar(line)<=pfx) {break}
if (substring(line, 1, pfx) != params$propertyPrefix) {break}
strArray=strsplit(substring(line, pfx+1, nchar(line)) ,"\t", fixed=TRUE)
prop<- strArray[[1]][1]
val<- strArray[[1]][2]
if (length(rpIn$name[rpIn$name==prop]) > 0 )
{
## dealing with dates is sometimes tricky. You want the value pushed to rpIn
## to be a string representing a date but in the default date format This data
## file uses a non-defualt date format that we explicitly convert to date using
## as.Date and a format string.
## Then convert it back to character using the default format.
if (rpIn$val2[rpIn$name==prop]=="java.util.Date")
{
val<-as.character(as.Date(val, "%b%d%y"))
}
rpIn$val1[rpIn$name==prop]<-val
fHasProps <- TRUE
}
}
if (fHasProps)
{
## write out the transformed run properties to the file that
## the assay framework will read in
write.table(rpIn, file=params$runPropsOutputPath, sep="\t", quote=FALSE
, na="" , row.names=FALSE, col.names=FALSE, append=FALSE)
}
return (params)
}
getLookupMap()
getLookupMap<- function(uniqueLookupValues, baseUrl, folderPath, schemaName,
queryName, keyField, displayField, otherColName=NULL, otherColValue=NULL)
{
inClauseVals = paste(uniqueLookupValues, collapse=";") #########
colfilt<-makeFilter(c(displayField, "EQUALS_ONE_OF", inClauseVals)) ## 4 ##
if (!is.null(otherColName)) #########
{
otherFilter=makeFilter(c(otherColName, "EQUALS", otherColValue))
colfilt = c(colfilt, otherFilter)
}
colsel<- paste(keyField, displayField, sep=",")
lookupMap <-labkey.selectRows(baseUrl=baseUrl, folderPath=folderPath,
schemaName=schemaName, queryName=queryName,
colSelect=colsel, colFilter=colfilt, showHidden=TRUE)
newLookups<- uniqueLookupValues[!(uniqueLookupValues %in% lookupMap[,2])]
if (length(newLookups)>0 && !is.na(newLookups[1]) )
{
## insert the lookup values that we haven't already seen before
newLookupsToInsert<- data.frame(lookupValue=newLookups, stringsAsFactors=FALSE)
colnames(newLookupsToInsert)<- displayField
if (!is.null(otherColName))
{
newLookupsToInsert<-cbind(newLookupsToInsert, otherColValue)
colnames(newLookupsToInsert)<- c(displayField, otherColName)
}
result<- labkey.insertRows(baseUrl=baseUrl, folderPath=folderPath,
schemaName=schemaName, queryName=queryName, toInsert= newLookupsToInsert)
lookupMap <-labkey.selectRows(baseUrl=baseUrl, folderPath=folderPath,
schemaName=schemaName, queryName=queryName,
colSelect=colsel, colFilter=colfilt, showHidden=TRUE)
}
colnames(lookupMap)<- c("RowId", "Name")
return(lookupMap)
}
doRunLoad()
doRunLoad<-function(params, inputColNames, outputColNames, lookupMap)
{
folder=params$containerPath
unlink(params$outputPath)
cIn <- file(params$inputPathUploadedFile, "r")
cOut<- file(params$outputPath , "w")
## write the column headers to the output file
headerDF<-data.frame(matrix(NA, nrow=0, ncol=length(outputColNames)))
colnames(headerDF)<- outputColNames
write.table(headerDF, file=cOut, sep="\t", quote=FALSE, row.names=FALSE, na="",
col.names=TRUE, append=FALSE)
# the fisrt read from the input file skips rows up to and including the header
skipCnt<-params$loaderColNamesRow
## read in chunks of batchSize, which are then transposed and written to the output file. #########
## blkStart is the 1-based index of the starting row of a chunk ## 5 ##
#########
blkStart <- skipCnt + 1
rowsToRead <- params$loaderBatchSize
while(rowsToRead > 0)
{
inputDF <- read.table(file=cIn, header = FALSE, sep = "\t", quote = "\"",
na.strings = "---", fill=TRUE, row.names=NULL,
stringsAsFactors = FALSE, check.names=FALSE,
col.names=inputColNames ,skip=skipCnt, nrows=rowsToRead)
cols<-colnames(inputDF)
if(NROW(inputDF) >0)
{
idVarName<-inputColNames[1]
df1 <- reshape(inputDF, direction="long", idvar=idVarName,,
v.names="Val",timevar="Name"
,times=cols[-1], varying=list(cols[-1]) ) #########
## 6 ##
df2<- merge(df1, lookupMap) #########
reshapedRows<- data.frame(cbind(df2[,idVarName], df2[,"RowId"],
df2[,"Val"], params$probeSourceId ), stringsAsFactors=FALSE)
reshapedRows[,2] <- as.integer(reshapedRows[,2])
reshapedRows[,4] <- as.integer(reshapedRows[,4])
nonEmptyRows<- !is.na(reshapedRows[,3])
reshapedRows<-reshapedRows[nonEmptyRows ,]
reshapedRows<- reshapedRows[ do.call(order, reshapedRows[1:2]), ]
colnames(reshapedRows)<- outputColNames
## need to double up the single quotes in the data
reshapedRows[,3]<-gsub("'", "''", reshapedRows[,3],fixed=TRUE)
write.table(reshapedRows, file=cOut, sep="\t", quote=TRUE, na="" ,
row.names=FALSE, col.names=FALSE, append=TRUE)
df1<-NULL
df2<-NULL
reshapedRows<-NULL
recordsToInsert<-NULL
}
if (NROW(inputDF)< rowsToRead)
{
##we've hit the end of the file, no more to read
rowsToRead <- 0
}
else
{
## now look where the next block will start, and read up to the end row
blkStart <- blkStart + rowsToRead
}
## skip rows only on the first read
skipCnt<-0
}
inputDF<-NULL
close(cIn)
close(cOut)
}
Transformation Scripts in Java
Overview
LabKey Server supports transformation scripts for assay data at upload time. This feature is primarily targeted for Perl or R scripts; however, the framework is general enough that any application that can be externally invoked can be run as well, including a Java program.
Java appeals to programmers who desire a stronger-typed language than most script-based languages. Most important, using a Java-based validator allows a developer to leverage the remote client API and take advantage of the classes available for assays, queries, and security.
This page outlines the steps required to configure and create a Java-based transform script. The ProgrammaticQCTest script, available in the BVT test, provides an example of a script that uses the remote client API.
Configure the Script Engine
In order to use a Java-based validation script, you will need to configure an external script engine to bind a file with the .jar extension to an engine implementation.
To do this:
- Go to the Admin Console for your site.
- Select the [views and scripting configuration] option.
- Create a new external script engine.
- Set up the script engine by filling in its required fields:
- File extension: jar
- Program path: (the absolute path to java.exe)
- Program command: -jar "${scriptFile}" "${runInfo}"
- scriptFile - The full path to the (processed and rewritten) transform script. This is usually in a temporary location the server manages.
- runInfo - The full path to the run properties file the server creates. For further info on this file, see the "Run Properties File" section of the Transformation Scripts documentation.
- srcDirectory - The original directory of the transform script (usually specified in the assay definition).
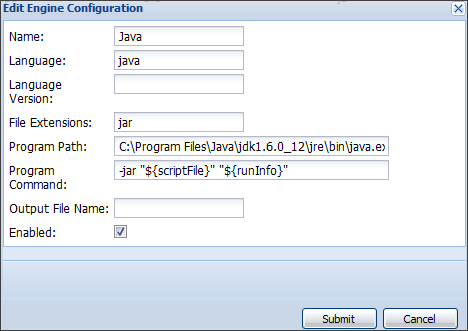
The program command configured above will invoke the java.exe application against a .jar file passing in the run properties file location as an argument to the java program. The run properties file contains information about the assay properties including the uploaded data and the location of the error file used to convey errors back to the server. Specific details about this file are contained in the data exchange specification for Programmatic QC.
Implement a Java Validator
The implementation of your java validator class must contain an entry point matching the following function signature:
public static void main(String[] args)
The location of the run properties file will be passed from the script engine configuration (described above) into your program as the first element of the args array.
The following code provides an example of a simple class that implements the entry point and handles any arguments passed in:
public class AssayValidator
{
private String _email;
private String _password;
private File _errorFile;
private Map<String, String> _runProperties;
private List<String> _errors = new ArrayList<String>();
private static final String HOST_NAME = "http://localhost:8080/labkey";
private static final String HOST = "localhost:8080";
public static void main(String[] args)
{
if (args.length != 1)
throw new IllegalArgumentException("Input data file not passed in");
File runProperties = new File(args[0]);
if (runProperties.exists())
{
AssayValidator qc = new AssayValidator();
qc.runQC(runProperties);
}
else
throw new IllegalArgumentException("Input data file does not exist");
}
Create a Jar File
Next, compile and jar your class files, including any dependencies your program may have. This will save you from having to add a classpath parameter in your engine command. Make sure that a ‘Main-Class’ attribute is added to your jar file manifest. This attribute points to the class that implements your program entry point.
Set Up Authentication for Remote APIs
Most of the remote APIs require login information in order to establish a connection to the server. Credentials can be hard-coded into your validation script or passed in on the command line. Alternatively, a .netrc file can be used to hold the credentials necesasry to login to the server. For further information, see: Create a .netrc or _netrc file.
The following sample code can be used to extract credentials from a .netrc file:
private void setCredentials(String host) throws IOException
{
NetrcFileParser parser = new NetrcFileParser();
NetrcFileParser.NetrcEntry entry = parser.getEntry(host);
if (null != entry)
{
_email = entry.getLogin();
_password = entry.getPassword();
}
}
Associate the Validator with an Assay Instance
Finally, the QC validator must be attached to an assay. To do this, you will need to editing the assay design and specify the absolute location of the .jar file you have created. The engine created earlier will bind the .jar extension to the java.exe command you have configured.
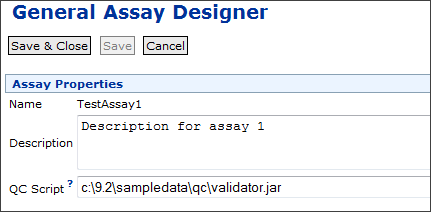
Transformation Scripts for Module-based Assays
<assay>
|_domains
|_views
|_scripts
|_config.xml
The scripts directory contains one or more script files; e.g., "validation.pl".
The order of script invocation can be specified in the config.xml file. See the <transformScripts> element. If scripts are not listed in the config.xml file, they will be executed in alphabetical order based on file name.
A script engine must defined for the appropriate type of script (for the example script named above, this would be a Perl engine). The rules for defining a script engine for module-based assays are the same as they are for Java-based assays.
When a new assay instance is created, you will notice that the script appears in the assay designer, but it is read-only (the path cannot be changed or removed). Just as for Java-defined assays, you will still see an additional text box where you can specify one or more additional scripts.
Run Properties Reference
There are standard default assay properties which apply to most assay types, as well as additional properties specific to the assay type. For example, NAb, Luminex, and ELISpot assays can include specimen, analyte, and antigen properties which correspond to locations on a plate associated with the assay instance.
The runProperties.tsv file also contains additional context information that the validation script might need, such as username, container path, assay instance name, assay id. Since the uploaded assay data will be written out to a file in TSV format, the runProperties.tsv also specifies the destination file's location.
Run Properties Format
The runProperties file has three (or four) tab-delimited columns in the following order:
- property name
- property value
- data type – The java class name of the property value (java.lang.String). This column may have a different meaning for properties like the run data, transformed data, or errors file. More information can be found in the property description below.
- transformed data location – The full path to the location where the transformed data are rewritten in order for the server to load them into the database.
Generic Assay Run Properties
| Property Name | Data Type | Property Description |
|---|---|---|
| assayId | String | The value entered in the Assay Id field of the run properties section. |
| assayName | String | The name of the assay design given when the new assay design was created. |
| assayType | String | The type of this assay design. (GenericAssay, Luminex, Microarray, etc.) |
| baseUrl | URL String | For example, http://localhost:8080/labkey |
| containerPath | String | The container location of the assay. (for example, /home/AssayTutorial) |
| errorsFile | Full Path | The full path to a .tsv file where any validation errors are written. See details below. |
| protocolDescription | String | The description of the assay definition when the new assay design was created. |
| protocolId | String | The ID of this assay definition. |
| protocolLsid | String | The assay definition LSID. |
| runComments | String | The value entered into the Comments field of the run properties section. |
| runDataUploadedFile | Full Path | The original data file that was selected by the user and uploaded to the server as part of an import process. This can be an Excel file, a tab-separated text file, or a comma-separated text file. |
| runDataFile | Full Path | The imported data file after the assay framework has attempted to convert the file to .tsv format and match its columns to the assay data result set definition. |
| transformedRunPropertiesFile | Full Path | File where the script writes out the updated values of batch- and run-level properties that are listed in the runProperties file. |
| userName | String | The user who created the assay design. |
| workingDir | String | The temp location that this script is executed in. (e.g. C:\AssayId_209\39\) |
errorsFile
Validation errors can be written to a TSV file as specified by full path with the errorsFile property. This output file is formatted with three columns:
- Type - "error" or "warn"
- Property - the name of the property raising the validation error
- Message - the actual error message
Additional Assay Specific Run Properties
ELISpot
| Property Name | Data Type | Property Description |
|---|---|---|
| sampleData | String | The path to a file that contains sample data written in a tab-delimited format. The file will contain all of the columns from the sample group section of the assay design. A wellgroup column will be written that corresponds to the well group name in the plate template associated with this assay instance. A row of data will be written for each well position in the plate template. |
| antigenData | String | The path to a file that contains antigen data written in a tab-delimited format. The file contains all of the columns from the antigen group section of the assay design. A wellgroup column corresponds to the well group name in the plate template associated with this assay instance. A row of data is written for each well position in the plate template. |
Luminex
| Property Name | Data Type | Property Description |
|---|---|---|
| Derivative | String | |
| Additive | String | |
| SpecimenType | String | |
| DateModified | Date | |
| ReplacesPreviousFile | Boolean | |
| TestDate | Date | |
| Conjugate | String | |
| Isotype | String |
NAb (TZM-bl Neutralizing Antibody) Assay
| Property Name | Data Type | Property Description |
|---|---|---|
| sampleData | String | The path to a file that contains sample data written in a tab-delimited format. The file contains all of the columns from the sample group section of the assay design. A wellgroup column corresponds to the well group name in the plate template associated with this assay instance. A row of data is written for each well position in the plate template. |
General Purpose Assay Type (GPAT)
| Property Name | Data Type | Property Description |
|---|---|---|
| severityLevel (reserved) | String | This is a property name used internally for error and warning handling. Do not define your own property with the same name in a GPAT assay. |
| maximumSeverity (reserved) | String | This is a property name reserved for use in error and warning handling. Do not define your own property with the same name in a GPAT assay. See Warnings in Tranformation Scripts for details. |
Transformation Script Substitution Syntax
| Script Syntax | Description | Substitution Value |
|---|---|---|
| ${runInfo} | File containing metadata about the run | Full path to the file on the local file system |
| ${srcDirectory} | Directory in which the script file is located | Full path to parent directory of the script |
| ${rLabkeySessionId} | Information about the current user's HTTP session | labkey.sessionCookieName = "COOKIE_NAME" labkey.sessionCookieContents = "USER_SESSION_ID" Note that this is multi-line. The cookie name is typically JSESSIONID, but is not in all cases.) |
| ${httpSessionId} | The current user's HTTP session ID | The string value of the session identifier, which can be used for authentication when calling back to the server for additional information |
| ${sessionCookieName} | The name of the session cookie | The string value of the cookie name, which can be used for authentication when calling back to the server for additional information. |
| ${baseServerURL} | The server's base URL and context path | The string of the base URL and context path. (ex. "http://localhost:8080/labkey") |
| ${containerPath} | The current container path | The string of the current container path. (ex. "/ProjectA/SubfolderB") |
Warnings in Tranformation Scripts
Enable Support for Warnings in a Transformation Script
To raise a warning from within your transformation script, set maximumSeverity to WARN within the transformedRunProperties file. To report an error, set maximumSeverity to ERROR. To display a specific message with either a warning or error, write the message to errors.html in the current directory. For example, this snippet from an R transformation script defines a warning and error handler:
# writes the maximumSeverity level to the transformRunProperties file and the error/warning message to the error.html file.
# LK server will read these files after execution to determine if an error or warning occurred and handle it appropriately
handleErrorsAndWarnings <- function()
{
if(run.error.level > 0)
{
fileConn<-file(trans.output.file);
if(run.error.level == 1)
{
writeLines(c(paste("maximumSeverity","WARN",sep="t")), fileConn);
}
else
{
writeLines(c(paste("maximumSeverity","ERROR",sep="t")), fileConn);
}
close(fileConn);
# This file gets read and displayed directly as warnings or errors, depending on maximumSeverity level.
if(!is.null(run.error.msg))
{
fileConn<-file("errors.html");
writeLines(run.error.msg, fileConn);
close(fileConn);
}
quit();
}
}
Click here to download a sample transformation script including this handler and other configuration required for warning reporting.
Workflow for Warnings from Transformation Scripts
When a warning is triggered during assay import, the user will see a screen similar to this with the option to Proceed or Cancel the import after examining the output files:
After examining the output and transformed data files, if the user clicks Proceed the transform script will be rerun and no warnings will be raised the on second pass. Quieting warnings on the approved import is handled using the value of an internal property called severityLevel in the run properties file. Errors will still be raised if necessary.
Priority of Errors and Warnings:
- 1. Script error (syntax, runtime, etc...) <- Error
- 2. Script returns a non-zero value <- Error
- 3. Script writes ERROR to maximumSeverity in the transformedRunProperties file <- Error
- If the script also writes a message to errors.html, it will be displayed, otherwise a server generated message will be shown.
- 4. Script writes WARN to maximumSeverity in the transformedRunProperties file <- Warning
- If the script also writes a message to errors.html, it will be displayed, otherwise a server generated message will be shown.
- The Proceed and Cancel buttons are shown, requiring a user selection to continue.
- 5. Script does not write a value to maximumSeverity in transformedRunProperties but does write a message to errors.html. This will be interpreted as an error.
Modules: ETLs
[ Video Update: ETL Enhancements in LabKey Server v15.1 ]
Extract-Transform-Load functionality lets you encapsulate some of the most common database tasks, especially (1) extracting data from a database, (2) transforming it, and finally (3) loading it into another database. LabKey Server ETL modules let you:
- Assemble data warehouses that integrate data from multiple data sources.
- Normalize data from different systems.
- Move data in scheduled increments.
- Log and audit migration processes.
- Tutorial: Extract-Transform-Load (ETL) - Develop an ETL module.
- ETL: Configuration and Schedules - Configure the refresh schedule.
- ETL: Column Mapping - Transform columns in an ETL process.
- ETL: Stored Procedures - Use a stored procedure as a transformation.
- ETL: SQL Scripts - Create table schemas with sql scripts.
- ETL: Logs and Error Handling - Logging and error reporting.
- ETL: Remote Connections - Extract data via a remote connection.
- ETL: Examples - Example ETL modules.
- ETL: Reference - File references, directory structure.
Related Topics
Tutorial: Extract-Transform-Load (ETL)
Data Warehouse
This tutorial shows you how to create a simple ETL as a starting point for further development.
As you go through the tutorial, imagine you are a researcher who wants to collect a group of participants for a research study. The participants must meet certain criteria to be included in the study, such as having a certain condition or diagnosis. You already have the following in place:
- You have a running installation of LabKey Server.
- You already have access to a large database of Demographic information of candidate participants. This database is continually being updated with new data and new candidates for your study.
- You have an empty table called "Patients" on your LabKey Server which is designed to hold the study candidates.
Tutorial Steps
- ETL Tutorial: Set Up - Set up a sample ETL workspace.
- ETL Tutorial: Run an ETL Process - Run an ETL.
- ETL Tutorial: Create a New ETL Process - Add a New ETL for a New Query.
First Step
ETL Tutorial: Set Up
- a basic workspace for working with ETL processes
- a working ETL module that can move data from the source database into the Patients table on your system.
Download
- Download the folder archive:
- ETLWorkspace.folder.zip (Don't unzip. This archive contains an ETL workspace and sample data to work with.)
- Download the server module:
Set Up ETL Workspace
In this step you will import a pre-configured workspace in which to develop ETL processes. (Note that there is nothing mandatory about the way this workspace has been put together -- your own ETL workspace may be different, depending on the needs of your project. This particular workspace has been configured especially for this tutorial as a shortcut to avoid many set up steps, steps such as connecting to source datasets, adding an empty dataset to use as the target of ETL scripts, and adding ETL-related web parts.)
- Go the LabKey Server Home project (or any project convenient for you).
- Create a subfolder of type Study to use as a workspace:
- Go to Admin > Folder > Management.
- Click Create Subfolder.
- On the Create Folder page, enter the Name "ETL Workspace".
- Under Folder Type, select Study.
- Click Next.
- On the Users/Permissions page, click Finish.
- Import ETLWorkspace.folder.zip into the folder:
- In the Study Overview panel, click Import Study.
- On the Folder Management page, confirm Local zip archive is selected and click Choose File.
- Select the folder archive that you have already downloaded: ETLWorkspace.folder.zip.
- Click Import Folder.
- When the import is complete, click ETL Workspace to see the workspace.
You now have a workspace where you can develop ETL scripts. It includes:
- A LabKey Study with various datasets to use as data sources
- An empty dataset named Patients to use as a target destination
- The ETLs tab provides an area to manage and run your ETL processes. Notice that this tab contains three panels/"web parts":
- Data Transforms shows the available ETL processes. Currently it is empty because there are none defined.
- The Patients dataset (the target dataset for the process) is displayed, also empty because no ETL process has been run yet. When you run an ETL process in the next step the the empty Patients dataset will begin to fill with data.
- The Demographics dataset (the source dataset for this tutorial) is displayed with more than 200 records.
Add the ETL Module
ETL processes are added to LabKey Server as part of a "module". Modules are packets of functionality that are easy to distribute to other LabKey Servers. Modules can contain a wide range of functionality, not just ETL-related functionality. For example, they can include HTML pages, SQL queries, R script reports, and more. Module resources are for the most part "file-based", that is, they contain files such as .HTML, .SQL, and .R files which are deployed to the server and surfaced in various places in the user interface where users can interact with them. For deployment to the server, the module files are zipped up into a .zip archive, which is renamed as a ".module" file. In this case, the module you will deploy contains two resources:
- An ETL configuration file (called "FemaleARV.xml") which defines how the ETL process works
- A SQL query which defines the source data for the ETL process
- If you are a developer working with the LabKey Server source code directly, then:
- Copy the file etlModule.module to the directory /build/deploy/modules and restart the server.
- If you are working with an installer-based version of LabKey Server, then:
- Copy the file etlModule.module to the directory LABKEY_HOME/externalModules and restart the server. (On Windows you can restart the server using the Services panel.)
- Enable the module in your workspace folder:
- In the "ETL Workspace" folder, go to Admin > Folder > Management. (You need to be logged in as an admin to complete this step.)
- Click the Folder Type tab.
- In the Modules list (on the right) place a checkmark next to ETLModule.
- Click Update Folder.
- The ETL script is now ready to run. Notice it has been added to the list under Data Transforms.
Start Over | Next Step
ETL Tutorial: Run an ETL Process
ETL User Interface
The web part Data Transforms lists all of the ETL processes that are available in the current folder. It lets you review current status at a glance, and run any transform manually or on a set schedule. You can also reset state after a test run.
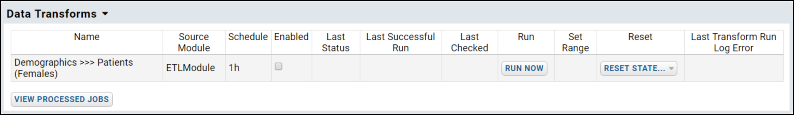
For details on the ETL user interface, see ETL: User Interface.
Run the ETL Process
- If necessary, click the ETLs tab, to return to the main page including the Data Transforms web part.
- Click Run Now for the "Demographics >>> Patients" row to transfer the data to the Patients table.
- You will be taken to the ETL Job page, which provides updates on the status of the running job.
- Refresh your browser until you see the Status field shows the value COMPLETE
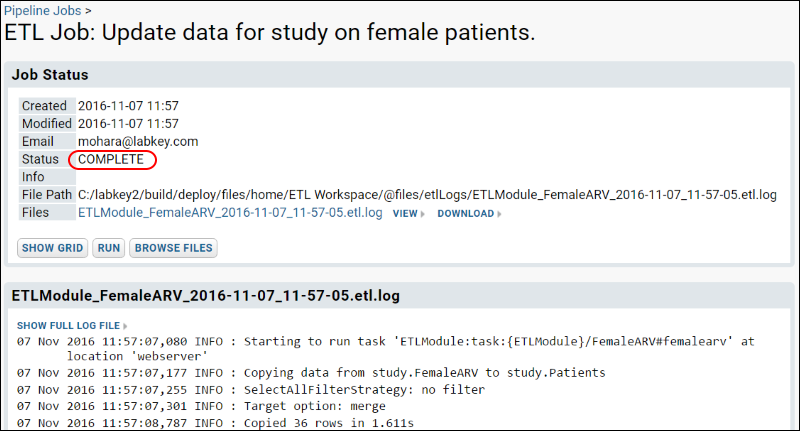
- Click the ETLs tab to see the records that have been added to the Patients table. Notice that 36 records (out of over 200 in the source Demographics query) have been copied into the Patients query. The ETL process is filtering to show female members of the ARV treatment group.
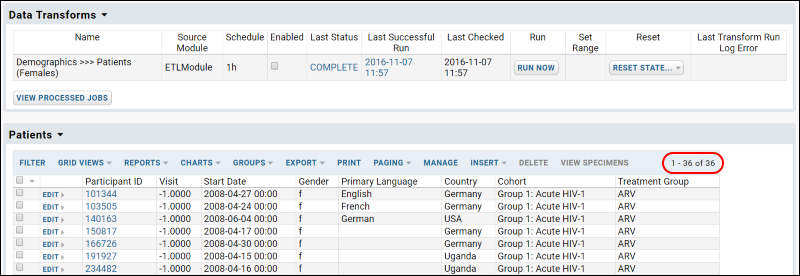
Experiment with ETL Runs
Now that you have a working ETL process, you can experiment with different scenarios.
- First, roll back the rows added to the target table (that is, delete the rows and return the target table to its original state) by selecting Reset State > Truncate and Reset.
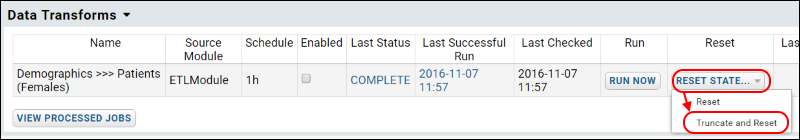
- Confirm the deletion in the popup window.
- Rerun the ETL process by clicking Run Now.
- The results are the same because we did not in fact change any source data yet. Next you can actually make some changes to show that they will be reflected.
- Edit the data in the source table Demographics:
- Click the ETLs tab.
- Scroll down to the Demographics dataset - remember this is our source data.
- Click Edit next to a record where the Gender is M and the Treatment Group is ARV. You could also apply column filters to find this set of records.
- Change the Gender to "F" and save.
- Rerun the ETL process by first selecting Reset > Truncate and Reset, then click Run Now.
- The resulting Patients table will now contain the additional matching row for a total count of 37 matching records.
Previous Step | Next Step
ETL Tutorial: Create a New ETL Process
To do this, we add a SQL query that returns a selection of records from the Demographics table, in particular all Male participants who are Natural Controllers.
And we'll create a new ETL process from scratch, drawing on the new SQL query.
Create a New Source Query
- Locate the source code for the ETL module. Depending on where you deployed it, go to either LABKEY_HOME/externalModules/etlmodule or build/deploy/modules/etlmodule.
- Go to the directory etlmodule/queries/study.
- In that directory, create a file named "MaleNC.sql".
- Open the file in a text editor and copy and paste the following code into the file:
SELECT Demographics.ParticipantId,
Demographics.StartDate,
Demographics.Gender,
Demographics.PrimaryLanguage,
Demographics.Country,
Demographics.Cohort,
Demographics.TreatmentGroup
FROM Demographics
WHERE Demographics.Gender = 'm' AND Demographics.TreatmentGroup = 'Natural Controller'
- Save the file.
- Restart the server.
Create a New ETL Process
ETL processes are defined by XML configuration files that specify the data source, the data target, and other properties. Here we create a new configuration that draws from the query we just created above.
- In the etlmodule/etls directory, create a new XML file called "MaleNC.xml".
- Copy the following into MaleNC.xml, and save.
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Demographics >>> Patients (Males)</name>
<description>Update data for study on male patients.</description>
<transforms>
<transform id="males">
<source schemaName="study" queryName="MaleNC"/>
<destination schemaName="study" queryName="Patients" targetOption="merge"/>
</transform>
</transforms>
<schedule>
<poll interval="1h"/>
</schedule>
</etl>
- Notice that this configuration file has our query (MaleNC) as its source, and the Patients query as its target.
- Refresh the browser. Notice that the system will add your new module to the server. Click Next if necessary to complete the update.
- In the "ETL Workspace" folder, notice our new ETL process is now listed in the Data Transforms web part.
Run the ETL Process
- Click Run Now next to the new process name. You will need to sign in to see this button.
- Refresh in the pipeline window until the job completes, then click the ETLs tab.
- New records will have been copied to the Patients table, making a total of 43 records (42 if you skipped the step of changing the gender of a participant in the previous tutorial step).
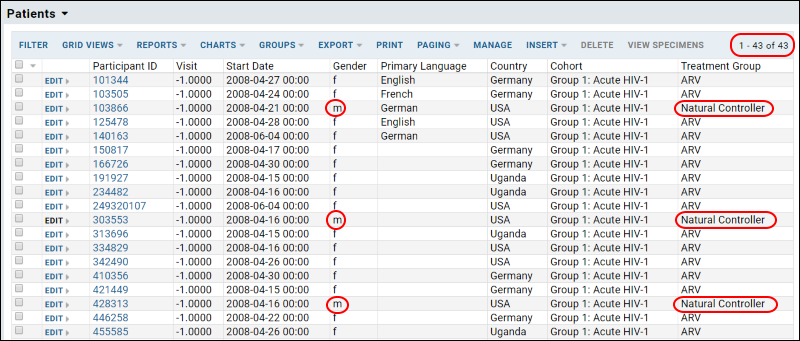
Previous Step
ETL: User Interface
ETL User Interface
The web part Data Transforms lists all of the ETL processes that are available in the current folder.
- Columns:
- Name - This column displays the name of the process.
- Source Module - This column tells you module where the configuration file resides.
- Schedule - This column shows you the reload schedule. In this case the ETL process is configured to run once every hour.
- Enabled - This checkbox controls whether the automated schedule is enabled: when unchecked, the ETL process must be run manually.
- Last Status, Successful Run, Checked - These columns record the latest run of the ETL process.
- Set Range - (Available only in devMode) The Set Range column is displayed only in dev mode and is intended for testing purposes during ETL module development. The Run button is only displayed for ETL processes with a filter strategy of RunFilterStrategy or ModifiedSinceFilterStrategy; the button is not displayed for the filter strategy SelectAllFilterStrategy. Click Run to set a date or row version window range to use for incremental ETL filters, overriding any persisted or initial values.
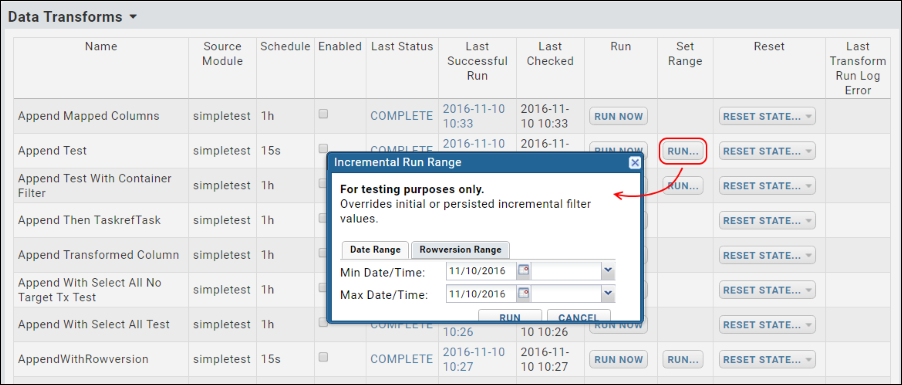
- Last Transform Run Log Error - Shows the last error logged, if any exists.
- Buttons:
- Run Now - This button immediately activates the ETL process.
- Reset State - This button returns the ETL process to its original state, deleting its internal history of which records are, and are not, up to date. There are two options:
- Reset
- Truncate and Reset
- View Processed Jobs - This button shows you a log of all previously run ETL jobs, and their status.
Run an ETL Process Manually
The Data Transforms web part lets you:
- Run jobs manually. (Click Run Now.)
- Enable/disable the recurring run schedule, if such a schedule has been configured in the ETL module. (Check or uncheck the column Enabled.)
- Reset state. (Select Reset State > Reset resets an ETL transform to its initial state, as if it has never been run.)
- See the latest error raised in the Last Transform Run Log Error column.
Cancel and Roll Back Jobs
While a job is running you can cancel and roll back the changes made by the current step by pressing the Cancel button.
The Cancel button is available on the Job Status panel for a particular job, as show below:
To roll back a run and delete the rows added to the target by the previous run, view the Data Transforms webpart, then select Reset State > Truncate and Reset. Note that rolling back an ETL which outputs to a file will have no effect, that is, the file will not be deleted or changed.
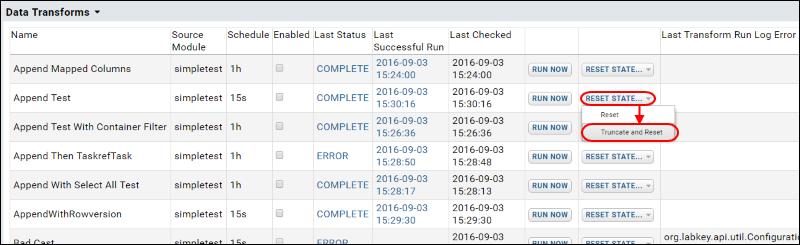
See Run History
The Data Transform Jobs web part provides a detailed history of all executed ETL runs, including the job name, the date and time when it was executed, the number of records processed, the amount of time spent to execute, and links to the log files.
To add this web part to your page, scroll down to the bottom of the page and click the dropdown <Select Web Part>, select Data Transform Jobs, and click Add. When added to the page, the web part appears with a different title: "Processed Data Transforms".
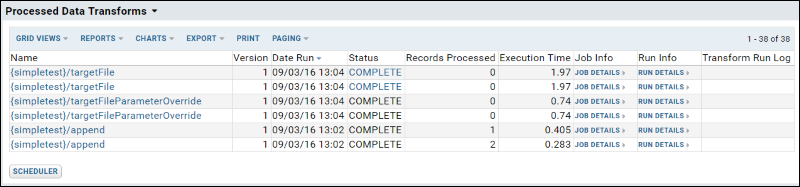
Click Run Details for fine-grained details about each run, including a graphical representation of the run.
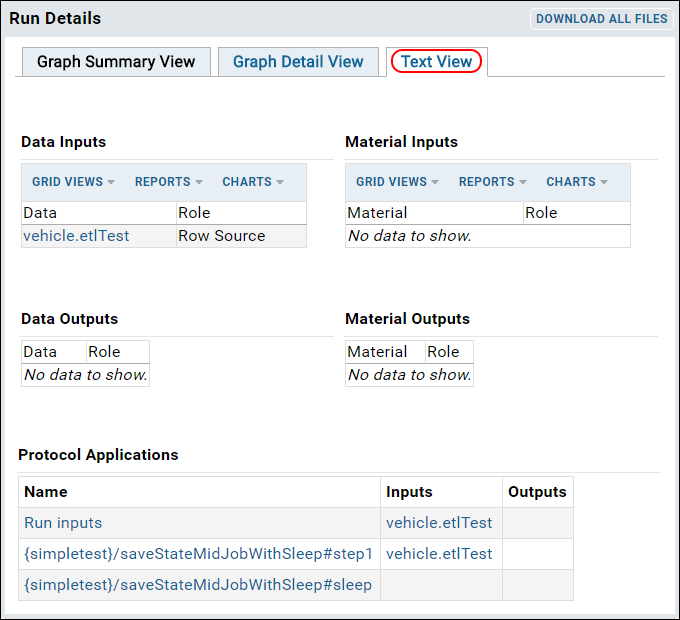
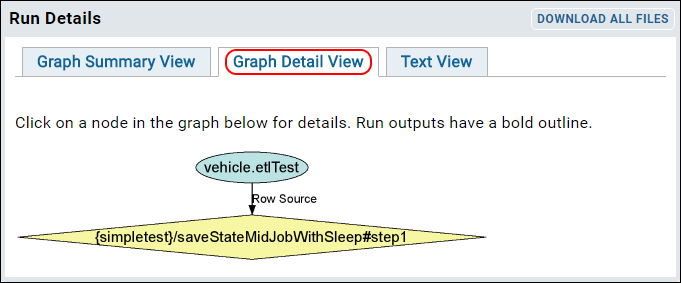
ETL: Configuration and Schedules
Schedules
You can set a polling schedule to check the source database for new data and automatically run the ETL process when new data is found. The schedule below checks every hour for new data:
<schedule><poll interval="1h" /></schedule>Another automatic scheduling option is to set a time each day to run the job.
<!-- run at 10:15 every day -->
<schedule><cron expression="0 15 10 ? * *"/></schedule>
<!-- run at 3:30am every day -->
<schedule><cron expression="0 30 3 * * ?"/></schedule>
Use an cron expression builder for the Quartz cron format, for example: http://www.cronmaker.com/
See quartz documentation for more examples.
Target Options
When the data is loaded into the destination database, there are three options for handling cases when the source query returns key values that already exist in the destination:
- Append: Appends new rows to the end of the existing table. Fails on duplicate primary key values.
- Merge: Merges data into the destination table. Matches primary key values to determine insert or update. Target tables must have a primary key.
- Truncate: Deletes the contents of the destination table before inserting the selected data.
<destination schemaName="vehicle" queryName="targetQuery" targetOption="merge" />
Note: Merge and truncate are only supported for datasets, not lists.
Filter Strategy
The filter strategy, defined in the incrementalFilter tag, is how the ETL process identifies new rows in the source database. The strategy allows a special value on the destination table to be compared to the source and only pulls over new rows based on that value. Using an incrementalFilter allows you to use the append option to add new rows to your target table and not accidentally run into any duplicate record conflicts. There are three options:
- SelectAllFilterStrategy: Apply no further filter to the source; simply transform/transfer all rows returned by the query.
- RunFilterStrategy: Check a specified column, typically an increasing integer column (e.g. Run ID), against a given or stored value. For instance, any rows with a higher value than when the ETL process was last run are transformed.
- ModifiedSinceFilterStrategy: Use a specified date/timestamp column (timeStampColumnName) to identify the new records. Rows changed since the last run will be transformed.
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="Date" />
Incremental Deletion of Target Rows
When incrementally deleting rows based on a selective filter strategy, use the element deletedRowsSource to correctly track the filtered values for deletion independently of the main query. Even if there are no new rows in the source query, any new records in the "deleteRowsSource" will still be found and deleted from the source. Using this method, the non-deleted rows will keep their row ids, maintaining any links to other objects in the target table.
File Targets
An ETL process can load data to a file, such as a comma separated file (CSV), instead of loading data into a database table. For example, the following ETL configuration element directs outputs to a tab separated file named "report.tsv". rowDelimiter and columnDelimiter are optional, if omitted you get a standard TSV file.
<destination type="file" dir="etlOut" fileBaseName="report" fileExtension="tsv" />
Transaction Options
Note that these settings risk leaving the destination or target table in an intermediate state if an error occurs during ETL processing.
ETL jobs are, by default, run as transactions. To turn off transactions when running an ETL process, set useTransaction to false on the destination, as shown below:
<destination schemaName="study" queryName="demographics" useTransaction="false" />
By default an single ETL job will be run as a single transaction, no matter how many rows are processed. You can change the default behavior by specifying that a new transaction be committed for every given number of rows processed. In the example below, a new transaction will be committed for every 500 rows processed:
<destination schemaName="study" queryName="demographics" bulkLoad="true" batchSize="500" />
Command Tasks
Once a command task has been registered in a pipeline task xml file, you can specify the task as an ETL step.
<transform id="ProcessingEngine" type="ExternalPipelineTask"
externalTaskId="org.labkey.api.pipeline.cmd.CommandTask:myEngineCommand"/>
ETL: Column Mapping
- Column Mapping
- Constants
- Creation and Modification Columns
- Data Integration Columns
- Transformation Java Classes
Column Mapping
If your source and target tables have different column names, you can configure a mapping between the columns, such that data from one column will be loaded into the mapped column, even if it has a different name. For example, suppose you are working with the following tables:
| Source Table Columns | Target Table Columns |
|---|---|
| ParticipantId | SubjectId |
| StartDate | Date |
| Gender | Sex |
| TreatmentGroup | Treatment |
| Cohort | Group |
Below we add a mapping such that data from "ParticipantId" is loaded into the column "SubjectId". Add column mappings to your ETL configuration using a <columnTransforms> element, with <column> elements to define each name mapping. For example:
<transform id="transform1">
<source schemaName="study" queryName="Participants"/>
<destination schemaName="study" queryName="Subjects" targetOption="merge">
<columnTransforms>
<column source="ParticipantId" target="SubjectId"/>
<column source="StartDate" target="Date"/>
<column source="Gender" target="Sex"/>
<column source="TreatmentGroup" target="Treatment"/>
<column source="Cohort" target="Group"/>
</columnTransforms>
</destination>
</transform>
Column mapping is supported for both query and file destinations. Mapping one source column onto many destination columns is not supported.
Container Columns
Container columns can be used to integrate data across different containers within LabKey Server. For example, data gathered in one project can be referenced from other locations as if it were available locally. However, ETL processes are limited to running within a single container. You cannot map a target container column to anything other than the container in which the ETL process is run.
Constants
To assign a constant value to a given target column, use a constant in your ETL configuration .xml file. For example, this sample would write "schema1.0" into the sourceVersion column of every row processed:
<constants>
<column name="sourceVersion" type="VARCHAR" value="schema1.0"/>
</constants>
If a column named "sourceVersion" exists in the source query, the constant value specified in your ETL xml file is used instead.
Constants can be set at both:
- The top level of your ETL xml: the constant is applied for every step in the ETL process.
- At an individual transform step level: the constant is only applied for that step and overrides any global constant that may have been set.
<destination schemaName="vehicle" queryName="etl_target">
<constants>
<column name="sourceVersion" type="VARCHAR" value="myStepValue"/>
</constants>
</destination>
Creation and Modification Columns
If the source table includes the following columns, they will be populated in the target table with the same names:
- EntityId
- Created
- CreatedBy
- Modified
- ModifiedBy
If no data is provided for these columns, they will be populated with the time and user information from the running of the ETL process.
DataIntegration Columns
Adding the following data integration ('di') columns to your target table will enable integration with other related data and log information.
| Column Name | PostresSQL Type | MS SQL Server Type | Notes |
|---|---|---|---|
| diTransformRunId | INT | INT | |
| diRowVersion | TIMESTAMP | DATETIME | |
| diModified | TIMESTAMP | DATETIME | Values here may be updated in later data mergers. |
| diModifiedBy | USERID | USERID | Values here may be updated in later data mergers. |
| diCreated | TIMESTAMP | DATETIME | Values here are set when the row is first inserted via a ETL process, and never updated afterwards |
| diCreatedBy | USERID | USERID | Values here are set when the row is first inserted via a ETL process, and never updated afterwards |
The value written to diTransformRunId will match the value written to the TransformRunId column in the table dataintegration.transformrun, indicating which ETL run was responsible for adding which rows of data to your target table.
Transformation Java Classes
The ETL pipeline allows Java developers to add a transformation java class to a particular column. This Java class can validate, transform or perform some other action on the data values in the column. For details and an example, see ETL: Examples
Reference
ETL: Queuing ETL Processes
Reference the ETL process you wish to queue up by module name and file name, using the pattern "{MODULE_NAME}/FILE_NAME". For example, to queue up the process MaleNC.xml in the module etlmodule, use the following:
<transforms>
...
<transform id="QueueTail" type="TaskrefTransformStep">
<taskref ref="org.labkey.di.steps.QueueJobTask">
<settings>
<setting name="transformId" value="{MODULE-NAME}/MaleNC"/>
</settings>
</taskref>
</transform>
...
</transforms>
An ETL process can also queue itself by omitting the <setting> element:
<transform id="requeueNlpTransfer" type="TaskrefTransformStep">
<taskref ref="org.labkey.di.steps.QueueJobTask"/>
</transform>
Handling Generated Files
If file outputs are involved (for example, if one ETL process outputs a file, and then queues another process that expects to use the file in a pipeline task), all ETL configurations in the chain must have the attribute loadReferencedFile="true” in order for the runs to link up properly.
<etl xmlns="http://labkey.org/etl/xml" loadReferencedFiles="true">
...
</etl>
Standalone vs. Component ETL Processes
ETL processes can be set as either "standalone" or "sub-component":
- Standalone ETL processes:
- Appear in the Data Transforms web part
- Can be run directly via the user or via another ETL
- Sub-Component ETL processes or tasks:
- Not shown in the Data Transforms web part
- Cannot be run directly by the user, but can be run only by another ETL process, as a sub-component of a wider job.
- Cannot be enabled or run directly via an API call.
<transform id="MySubComponent" standalone="false">
...
</transform>
ETL: Stored Procedures
Stored Procedures as Source Queries
Instead of extracting data directly from a source query and loading it into a target query, an ETL process can call one or more stored procedures that themselves move data from the source to the target (or the procedures can transform the data in some other way). For example, the following ETL process runs a stored procedure to populate the Patients table.
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Populate Patient Table</name>
<description>Populate Patients table with calculated and converted values.</description>
<transforms>
<transform id="ExtendedPatients" type="StoredProcedure">
<description>Calculates date of death or last contact for a patient, and patient ages at events of interest</description>
<procedure schemaName="patient" procedureName="PopulateExtendedPatients" useTransaction="true">
</procedure>
</transform>
</transforms>
<!-- run at 3:30am every day -->
<schedule><cron expression="0 30 3 * * ?"/></schedule>
</etl>
Special Behavior for Different Database Implementations
ETL: Stored Procedures in MS SQL Server
Example - Normalize Data
The following ETL process uses the stored procedure normalizePatientData to modify the source data.
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Target #1 (Normalize Gender Values - Stored Procedure)</name>
<description>Runs a stored procedure.</description>
<transforms>
<transform id="storedproc" type="StoredProcedure">
<description>Runs a stored procedure to normalize values in the Gender column.</description>
<procedure schemaName="target1" procedureName="normalizePatientData">
</procedure>
</transform>
</transforms>
</etl>
The stored procedure is shown below.
CREATE procedure [target1].[normalizePatientData] (@transformRunId integer)
as
begin
UPDATE Patients SET Gender='Female' WHERE (Gender='f' OR Gender='F');
UPDATE Patients SET Gender='Male' WHERE (Gender='m' OR Gender='M');
end
GO
Parameters
The <procedure> element can have <parameter> child elements that specify the initial seed values passed in as input/output parameters. Note that The "@" sign prefix for parameter names in the ETL xml configuration is optional.
<procedure … >
<parameter name="@param1" value="100" override="false"/>
<parameter name="@param2" value="200" override="false"/>
</procedure>
The output values of all input/output parameters are persisted in the database, and are used as input values for the next pass. These values take precedence over the initial seed values specified in the xml file. To reset and force the use of the value from the xml file, set the optional override attribute to "true".
<procedure schemaName="external" procedureName="etlTestRunBased">
<parameter name="@femaleGenderName" value="Female" override="false"/>
<parameter name="@maleGenderName" value="Male" override="false"/>
</procedure>
CREATE procedure [target1].[normalizePatientData] (@transformRunId integer,
@maleGenderName VARCHAR(25),
@femaleGenderName VARCHAR(25))
as
begin
UPDATE Patients SET Gender=@femaleGenderName WHERE (Gender='f' OR Gender='F');
UPDATE Patients SET Gender=@maleGenderName WHERE (Gender='m' OR Gender='M');
end
GO
Parameters - Special Processing
The following parameters are given special processing.
| Name | Direction | Datatype | Notes |
|---|---|---|---|
| @transformRunId | Input | int | Assigned the value of the current transform run id. |
| @filterRunId | Input or Input/Output | int | For RunFilterStrategy, assigned the value of the new transfer/transform to find records for. This is identical to SimpleQueryTransformStep’s processing. For any other filter strategy, this parameter is available and persisted for stored procedure to use otherwise. On first run, will be set to -1. |
| @filterStartTimestamp | Input or Input/Output | datetime | For ModifiedSinceFilterStrategy with a source query, this is populated with the IncrementalStartTimestamp value to use for filtering. This is the same as SimpleQueryTransformStep. For any other filter strategy, this parameter is available and persisted for stored procedure to use otherwise. On first run, will be set to NULL. |
| @filterEndTimestamp | Input or Input/Output | datetime | For ModifiedSinceFilterStrategy with a source query, this is populated with the IncrementalEndTimestamp value to use for filtering. This is the same as SimpleQueryTransformStep. For any other filter strategy, this parameter is available and persisted for stored procedure to use otherwise. On first run, will be set to NULL. |
| @containerId | Input | GUID/Entity ID | If present, will always be set to the id for the container in which the job is run. |
| @rowsInserted | Input/Output | int | Should be set within the stored procedure, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| @rowsDeleted | Input/Output | int | Should be set within the stored procedure, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| @rowsModified | Input/Output | int | Should be set within the stored procedure, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| @returnMsg | Input/Output | varchar | If output value is not empty or null, the string value will be written into the output log. |
| @debug | Input | bit | Convenience to specify any special debug processing within the stored procedure. May consider setting this automatically from the Verbose flag. |
| Return Code | special | int | All stored procedures must return an integer value on exit. “0” indicates correct processing. Any other value will indicate an error condition and the run will be aborted. |
To write to the ETL log file, use a 'print' statement inside the procedure.
Log Rows Modified
Use special parameters to log the number of rows inserted, changed, etc. as follows:
CREATE procedure [target1].[normalizePatientData] (@transformRunId integer
, @parm1 varchar(25) OUTPUT
, @gender varchar(25) OUTPUT
, @rowsInserted integer OUTPUT
, @rowCount integer OUTPUT
, @rowsDeleted integer OUTPUT
, @rowsModified integer OUTPUT
, @filterStartTimestamp datetime OUTPUT)
as
begin
SET @rowsModified = 0
UPDATE Patients SET Gender='Female' WHERE (Gender='f' OR Gender='F');
SET @rowsModified = @@ROWCOUNT
UPDATE Patients SET Gender='Male' WHERE (Gender='m' OR Gender='M');
SET @rowsModified += @@ROWCOUNT
end
Optional Source
An optional source must be used in combination with the RunFilterStrategy or ModifiedSinceFilterStrategy filter strategies.
<transforms>
<transform id="storedproc" type="StoredProcedure">
<description>
Runs a stored procedure to normalize values in the Gender column.
</description>
<!-- Optional source element -->
<!-- <source schemaName="study" queryName="PatientsWarehouse"/> -->
<procedure schemaName="target1" procedureName="normalizePatientData">
</procedure>
</transform>
</transforms>
Transactions
By default all stored procedures are wrapped as transactions, so that if any part of the procedure fails, any changes already made are rolled back. For debugging purposed, turn off the transaction wrapper setting useTransaction to "false":
<procedure schemaName="target1" procedureName="normalizePatientData" useTransaction="false">
</procedure>
ETL: Functions in PostgreSQL
To call a PostgreSQL function from an ETL process, refer to the function in a transform element of the ETL configuration file. For example, the following ETL process calls "postgresFunction" in the patient schema.
ETL XML Configuration File
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Stored Proc Normal Operation</name>
<description>Normal operation</description>
<transforms>
<transform id="callfunction" type="StoredProcedure">
<procedure schemaName="patient" procedureName="postgresFunction" useTransaction="false">
<parameter name="inoutparam" value="before"/>
</procedure>
</transform>
</transforms>
</etl>
Function and Parameter Requirements
PostgreSQL functions called by an ETL process must meet the following requirements:
- The Postgres function must be of return type record.
- Parameter names, including the Special Processing parameters (see table below), are case-insensitive.
- There can be an arbitrary number of custom INPUT and/or INPUT/OUTPUT parameters defined for the function.
- There can be at most one pure OUTPUT parameter. This OUTPUT parameter must be named "return_status" and must be of type INTEGER. If present, the return_status parameter must be assigned a value of 0 for successful operation. Values > 0 are interpreted as error conditions.
- Function overloading of differing parameter counts is not currently supported. There can be only one function (procedure) in the Postgres database with the given schema & name combination.
- Optional parameters in PostgreSQL are not currently supported. An ETL process using a given function must provide a value for every custom parameter defined in the function.
- Postgres does not have a "print" statement. Writing to the ETL log can be accomplished with a "RAISE NOTICE" statement, for example:
RAISE NOTICE '%', 'Test print statement logging';
- The "@" sign prefix for parameter names in the ETL configuration xml is optional (for both SQL Server and Postgres). When IN/OUT parameters are persisted in the dataintegration.transformConfiguration.transformState field, their names are consistent with their native dialect (an "@" prefix for SQL Server, no prefix for Postgres).
Parameters - Special Processing
The following parameters are given special processing.
Note that the output values of INOUT's are persisted to be used as inputs on the next run.
| Name | Direction | Datatype | Notes |
|---|---|---|---|
| transformRunId | Input | int | Assigned the value of the current transform run id. |
| filterRunId | Input or Input/Output | int | For RunFilterStrategy, assigned the value of the new transfer/transform to find records for. This is identical to SimpleQueryTransformStep's processing. For any other filter strategy, this parameter is available and persisted for functions to use otherwise. On first run, will be set to -1. |
| filterStartTimestamp | Input or Input/Output | datetime | For ModifiedSinceFilterStrategy with a source query, this is populated with the IncrementalStartTimestamp value to use for filtering. This is the same as SimpleQueryTransformStep. For any other filter strategy, this parameter is available and persisted for functions to use otherwise. On first run, will be set to NULL. |
| filterEndTimestamp | Input or Input/Output | datetime | For ModifiedSinceFilterStrategy with a source query, this is populated with the IncrementalEndTimestamp value to use for filtering. This is the same as SimpleQueryTransformStep. For any other filter strategy, this parameter is available and persisted for functions to use otherwise. On first run, will be set to NULL. |
| containerId | Input | GUID/Entity ID | If present, will always be set to the id for the container in which the job is run. |
| rowsInserted | Input/Output | int | Should be set within the function, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| rowsDeleted | Input/Output | int | Should be set within the functions, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| rowsModified | Input/Output | int | Should be set within the functions, and will be recorded as for SimpleQueryTransformStep. Initialized to -1. Note: The TransformRun.RecordCount is the sum of rows inserted, deleted, and modified. |
| returnMsg | Input/Output | varchar | If output value is not empty or null, the string value will be written into the output log. |
| debug | Input | bit | Convenience to specify any special debug processing within the stored procedure. |
| return_status | special | int | All functions must return an integer value on exit. “0” indicates correct processing. Any other value will indicate an error condition and the run will be aborted. |
Example Postgres Function
CREATE OR REPLACE FUNCTION patient.postgresFunction
(IN transformrunid integer
, INOUT rowsinserted integer DEFAULT 0
, INOUT rowsdeleted integer DEFAULT 0
, INOUT rowsmodified integer DEFAULT 0
, INOUT returnmsg character varying DEFAULT 'default message'::character varying
, IN filterrunid integer DEFAULT NULL::integer
, INOUT filterstarttimestamp timestamp without time zone DEFAULT NULL::timestamp without time zone
, INOUT filterendtimestamp timestamp without time zone DEFAULT NULL::timestamp without time zone
, INOUT runcount integer DEFAULT 1
, INOUT inoutparam character varying DEFAULT ''::character varying
, OUT return_status integer)
RETURNS record AS
$BODY$
BEGIN
/*
*
* Function logic here
*
*/
RETURN;
END;
$BODY$
LANGUAGE plpgsql;
ETL: Check For Work From a Stored Procedure
<transform id="checkToRun" type="StoredProcedure">
<procedure schemaName="patient" procedureName="workcheck" useTransaction="false">
<parameter name="StagingControl" value="1" noWorkValue="-1"/>
</procedure>
</transform>
<transform id="queuedJob">
<source schemaName="patient_source" queryName="etl_source" />
<destination schemaName="patient_target" queryName="Patients" targetOption="merge"/>
</transform>
The noWorkValue can either be a hard-coded string (for example, "-1", shown above), or you can use a substitution syntax to indicate a comparison should be against the input value of a certain parameter.
For example, the following parameter indicates there is no work for the ETL job if the output batchId is the same as the output parameter persisted from the previous run.
<parameter name="batchId" noWorkValue="${batchId}"/>
Example
In the ETL transform below, the gating procedure checks if there is a new ClientStagingControlID to process. If there is, the ETL job goes into the queue. When the job starts, the procedure is run again in the normal job context; the new ClientStagingControlID is returned again. The second time around, the output value is persisted into the global space, so further procedures can use the new value. Because the gating procedure is run twice, don’t use this with stored procedures that have other data manipulation effects! There can be multiple gating procedures, and each procedure can have multiple gating params, but during the check for work, modified global output param values are not shared between procedures.
<transform id="CheckForWork" type="StoredProcedure">
<description>Check for new batch</description>
<procedure schemaName="patient" procedureName="GetNextClientStagingControlID">
<parameter name="ClientStagingControlID" value="-1" scope="global" noWorkValue="${ClientStagingControlID}"/>
<parameter name="ClientSystem" value="LabKey-nlp-01" scope="global"/>
<parameter name="StagedTable" value="PathOBRX" scope="global"/>
</procedure>
</transform>
ETL: SQL Scripts
Directory Structure
LabKey Server will automatically run SQL scripts that are packaged inside your module in the following directory structure:
MODULE_NAME config etls queries schemas dbscripts postgres SCRIPT_NAME.sql - Script for PostgreSQL. mssql SCRIPT_NAME.sql - Script for MS SQL Server.
SQL Script Names
Script names are formed from three components: (1) schema name, (2) previous module version, and (3) current module version, according to the following pattern:
SCHEMA-PREVIOUSVERSION-CURRENTVERSION.sql
where SCHEMA is the name of the schema to be generated by the script.
For an initially deployed module that hasn't existed on the server previously, an example script name would be:
patientSchema-0.0-1.0.sql
For more details on naming scripts, especially naming upgrade scripts, see Modules: SQL Scripts.
Schema XML File
LabKey will generate an XML schema file for a table schema by visiting a magic URL of the form:
http://<server>/labkey/admin/getSchemaXmlDoc.view?dbSchema=<schema-name>
Examples
This script creates a simple table and a stored procedure for MS SQL Server dialect.
CREATE SCHEMA target1;
GO
CREATE procedure [target1].[normalizePatientData] (@transformRunId integer)
as
begin
UPDATE Patients SET Gender='Female' WHERE (Gender='f' OR Gender='F');
UPDATE Patients SET Gender='Male' WHERE (Gender='m' OR Gender='M');
end
GO
CREATE TABLE target1.Patients
(
RowId INT IDENTITY(1,1) NOT NULL,
Container ENTITYID NOT NULL,
CreatedBy USERID NOT NULL,
Created DATETIME NOT NULL,
ModifiedBy USERID NOT NULL,
Modified DATETIME NOT NULL,
PatientId INT NOT NULL,
Date DATETIME NOT NULL,
LastName VARCHAR(30),
FirstName VARCHAR(30),
MiddleName VARCHAR(30),
DateVisit DATETIME,
Gender VARCHAR(30),
PrimaryLanguage VARCHAR(30),
Email VARCHAR(30),
Address VARCHAR(30),
City VARCHAR(30),
State VARCHAR(30),
ZIP VARCHAR(30),
Diagnosis VARCHAR(30),
CONSTRAINT PatientId PRIMARY KEY (RowId)
);
These scripts are in Postgres SQL dialect.
---------------
-- schema1 --
---------------
DROP SCHEMA schema1 CASCADE;
CREATE SCHEMA schema1;
CREATE TABLE schema1.patients
(
patientid character varying(32),
date timestamp without time zone,
startdate timestamp without time zone,
country character varying(4000),
language character varying(4000),
gender character varying(4000),
treatmentgroup character varying(4000),
status character varying(4000),
comments character varying(4000),
CONSTRAINT patients_pk PRIMARY KEY (patientid)
);
CREATE OR REPLACE FUNCTION changecase(searchtext varchar(100), replacetext varchar(100)) RETURNS integer AS $$
UPDATE schema1.patients
SET gender = replacetext
WHERE gender = searchtext;
SELECT 1;
$$ LANGUAGE SQL;
Related Topics
- Tutorial: Extract-Transform-Load (ETL) - Other SQL scripts are included in the ETL samples at patientetl/schemas/dbscripts.
- Modules: SQL Scripts - Details on SQL scripts in modules.
ETL: Remote Connections
To set up a remote connection, see Manage Remote Connections.
To configure an ETL process to utilize a remote connection, specify the transform type and the remoteSource as shown below:
<transform type="RemoteQueryTransformStep" id="step1">
<source remoteSource="EtlTest_RemoteConnection" schemaName="study" queryName="etl source" />
...
</transform>
A sample ETL configuration file is shown below:
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Remote Test</name>
<description>append rows from "remote" etl_source to etl_target</description>
<transforms>
<transform type="RemoteQueryTransformStep" id="step1">
<description>Copy to target</description>
<source remoteSource="EtlTest_RemoteConnection" schemaName="study" queryName="etl source" />
<destination schemaName="study" queryName="etl target" targetOption="truncate"/>
</transform>
</transforms>
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="modified" />
</etl>
Related Topics
ETL: Logs and Error Handling
Logging
Messages and/or errors inside an ETL job are written to a log file named for that job, located at
LABKEY_HOME/files/PROJECT/FOLDER_PATH/@files/etlLogs/ETLNAME_DATE.etl.log
for example:
C:/labkey/files/MyProject/MyFolder/@files/etlLogs/myetl_2015-07-06_15-04-27.etl.log
Attempted/completed jobs and log locations are recorded in the table dataIntegration.TransformRun. For details on this table, see ETL: User Interface.
Log locations are also available from the Data Transform Jobs web part (named Processed Data Transforms by default). For the ETL job in question, click Job Details.
File Path shows the log location.
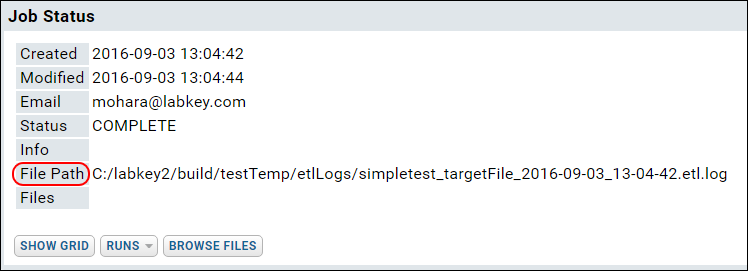
ETL processes check for work (= new data in the source) before running a job. Log files are only created when there is work. If, after checking for work, a job then runs, errors/exceptions throw a PipelineJobException. The UI shows only the error message; the log captures the stacktrace.
XSD/XML-related errors are written to the labkey.log file, located at TOMCAT_HOME/logs/labkey.log.
DataIntegration Columns
To record a connection between a log entry and rows of data in the target table, add the 'di' columns listed here to your target table.
Error Handling
If there were errors during the transform step of the ETL process, you will see the latest error in the Transform Run Log column.
- An error on any transform step within a job aborts the entire job. “Success” in the log is only reported if all steps were successful with no error.
- If the number of steps in a given ETL process has changed since the first time it was run in a given environment, the log will contain a number of DEBUG messages of the form: “Wrong number of steps in existing protocol”. This is an informational message and does not indicate anything was wrong with the job.
- Filter Strategy errors. A “Data Truncation” error may mean that the xml filename is too long. Current limit is module name length + filename length - 1, must be <= 100 characters.
- Stored Procedure errors. “Print” statements in the procedure appear as DEBUG messages in the log. Procedures should return 0 on successful completion. A return code > 0 is an error and aborts job.
- Known issue: When the @filterRunId parameter is specified in a stored procedure, a default value must be set. Use NULL or -1 as the default.
ETL: All Jobs History
The history includes the name of the job, the folder it was run in, the date and time it was run, and other information. Links to detailed views of each job are provided.
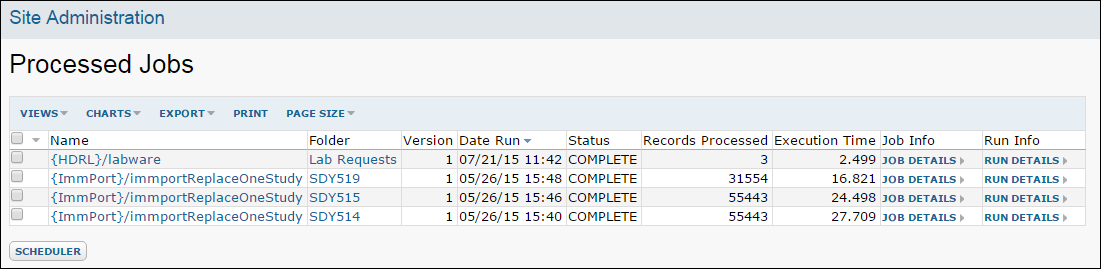
ETL: Examples
- Interval - 1 Hour
- Interval - 5 Minutes
- Cron - 1 Hour
- Merge
- Merge By Run ID
- Merge with Alternate Key
- Append with Two Targets
- Truncate
- Passing Parameters to a SQL Query
- Truncate the Target Query
- Java Transforms
Interval - 1 Hour
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Patient - Merge</name>
<description>Merges patient data to the target query.</description>
<transforms>
<transform id="1hour">
<source schemaName="external" queryName="etl_source" />
<destination schemaName="patient" queryName="etl_target" targetOption="merge"/>
</transform>
</transforms>
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="modified" />
<schedule><poll interval="1h"></poll></schedule>
</etl>
Interval - 5 Minutes
<schedule><poll interval="5m" /></schedule>
Cron - 1 Hour
Check at midnight every day.
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Cron on the hour</name>
<transforms>
<transform id="eachHour">
<description>valid</description>
<source schemaName="external" queryName="etl_source" />
<destination schemaName="patient" queryName="etl_target" targetOption="merge"/>
</transform>
</transforms>
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="modified" />
<schedule><cron expression="0 0 * * * ?" /></schedule>
</etl>
Merge
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Merge</name>
<description>Merge rows from etl_source to etl_target.</description>
<transforms>
<transform id="merge">
<description>Merge to target.</description>
<source schemaName="external" queryName="etl_source" />
<destination schemaName="patient" queryName="etl_target" targetOption="merge"/>
</transform>
</transforms>
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="modified" />
</etl>
Merge by Run ID
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>MergeByRunId</name>
<description>Merge by run id.</description>
<transforms>
<transform id="step1">
<description>Copy to target</description>
<source schemaName="patient" queryName="etlsource" />
<destination schemaName="target" queryName="etltarget" />
</transform>
</transforms>
<incrementalFilter className="RunFilterStrategy" runTableSchema="patient"
runTable="Transfer" pkColumnName="Rowid" fkColumnName="TransformRun" />
<schedule>
<poll interval="15s" />
</schedule>
</etl>
Merge with Alternate Key
Specify an alternate key to use for merging when the primary key is not suitable, i.e. would cause duplicates or orphaned data.
<destination schemaName="vehicle" queryName="etl_target2" targetOption="merge">
<alternateKeys>
<!-- The pk of the target table is the "rowId" column. Use "id" as an alternate match key -->
<column name="id"/>
</alternateKeys>
</destination>
Append with Two Targets
For example, you might want to ensure that a given stored procedure is executed (step1) before loading the data into the destination (step2).
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Append</name>
<description>append rows from etl_source to etl_target and etl_target2</description>
<transforms>
<transform id="step1">
<description>Copy to target</description>
<source schemaName="external" queryName="etl_source" timestampcolumnname="modfiied" />
<destination schemaName="patient" queryName="etl_target" />
</transform>
<transform id="step2">
<description>Copy to target two</description>
<source schemaName="external" queryName="etl_source" />
<destination schemaName="patient" queryName="etl_target2" />
</transform>
</transforms>
<incrementalFilter className="SelectAllFilterStrategy"/>
</etl>
Truncate
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Truncate</name>
<description>Clear target and append rows from etl_source.</description>
<transforms>
<transform id="step1">
<description>Copy to target</description>
<source schemaName="patient" queryName="etl_source" />
<destination schemaName="patient" queryName="etl_target" targetOption="truncate"/>
</transform>
</transforms>
<incrementalFilter className="ModifiedSinceFilterStrategy" timestampColumnName="modified" />
<schedule>
<poll interval="15s" />
</schedule>
</etl>
Passing Parameters to a SQL Query
The following ETL process passes parameters (MinTemp=99 and MinWeight=150) into its source query (a parameterized query).
<?xml version="1.0" encoding="UTF-8" ?>
<etl xmlns="http://labkey.org/etl/xml">
<name>PatientsToTreated</name>
<description>Transfers from the Patients table to the Treated table.</description>
<transforms>
<transform id="step1">
<description>Patients to Treated Table</description>
<source queryName="Patients" schemaName="study"/>
<destination schemaName="study" queryName="Treated"/>
</transform>
</transforms>
<parameters>
<parameter name="MinTemp" value="99" type="DECIMAL" />
<parameter name="MinWeight" value="150" type="DECIMAL" />
</parameters>
<schedule>
<poll interval="1h"/>
</schedule>
</etl>
Truncate the Target Query
The following truncates the target table, without copying any data from a source query. Note the lack of a <source> element.
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Truncate Patients Table</name>
<description>Update data for study on male patients.</description>
<transforms>
<transform id="trunc">
<destination schemaName="study" queryName="Patients" targetOption="truncate"/>
</transform>
</transforms>
<schedule>
<poll interval="1h"/>
</schedule>
</etl>
Java Transforms
Java developers can add a Java class to handle the transformation step of an ETL process. The column to be transformed points to the Java class as follows:<columnTransforms>
<column source="columnToTransform" transformClass="org.labkey.di.columnTransforms.MyJavaClass"/>
</columnTransforms>
The Java class receives the values of the column one row at a time. The Java class can validate, transform or perform some other action on these values. What is returned from the doTransform method of this class is what gets inserted into the target table. See below for an example implementation. Also see the ColumnTransform interface for available setters, getters, and methods.
The ETL source below uses the Java class org.labkey.di.columnTransforms.TestColumnTransform to apply changes to data in the "name" column.
ETL.xml
<?xml version="1.0" encoding="UTF-8"?>
<etl xmlns="http://labkey.org/etl/xml">
<name>Append Transformed Column</name>
<description>Append rows from etl_source to etl_target, applying column transformation using a Java class.</description>
<transforms>
<transform id="step1" type="org.labkey.di.pipeline.TransformTask">
<description>Copy to target</description>
<source schemaName="vehicle" queryName="etl_source" />
<destination schemaName="vehicle" queryName="etl_target">
<columnTransforms>
<column source="name" transformClass="org.labkey.di.columnTransforms.TestColumnTransform"/>
</columnTransforms>
<constants>
<column name="myConstant" type="varchar" value="aConstantValue"/>
</constants>
</destination>
</transform>
</transforms>
<incrementalFilter className="SelectAllFilterStrategy" />
</etl>
The Java class below is used by the ETL process to apply transformations to the supplied column, in this case the "name" column.
TestColumnTransform.java
package org.labkey.di.columnTransforms;
import org.labkey.api.di.columnTransform.AbstractColumnTransform;
/**
* An example of Java implementing a transform step.
* Prepends the value of the "id" column of the source query
* to the value of the source column specified in the ETL configuration xml,
* then appends the value of the "myConstant" constant set in the xml.
*/
public class TestColumnTransform extends AbstractColumnTransform
{
@Override
protected Object doTransform(Object inputValue)
{
Object prefix = getInputValue("id");
String prefixStr = null == prefix ? "" : prefix.toString();
return prefixStr + "_" + inputValue + "_" + getConstant("myConstant");
}
}
ETL: Reference
Directory Structure of an ETL Module
The directory structure for an ETL module is shown below. Note that the "queries" and "schemas" directories are optional, and not required for ETL functionality. Items shown in lowercase are literal values that should be preserved in the directory structure. Items shown in uppercase should be replaced with values that reflect the nature of your project.
MODULE_NAME
├───etls
│ ETL1.xml
│ ETL2.xml
│
├───queries
│ └───SCHEMA_NAME
│ QUERY_NAME.sql
│
└───schemas
│ SCHEMA_NAME.xml
│
└───dbscripts
├───postgresql
│ SCHEMA_NAME-X.XX-Y.YY.sql
│
└───sqlserver
SCHEMA_NAME-X.XX-Y.YY.sql
Files and Directories
- ETL1.xml - The main config file for an ETL process. Defines the sources, targets, transformations, and schedules for the transfers. Any number of ETL processes and tasks can be added. For examples see ETL: Examples.
- QUERY_NAME.sql - SQL queries for data sources and targets.
- schemas - Optional database schemas. Optional sql scripts for bootstrapping a target database.
ETL Configuration Reference
For details see ETL XML Reference.
Reference Links
- etl.xml Reference - xml reference docs for ETL config files.
- etl.xsd - The XSD schema file on which the XML config files are based.
Modules: Java
Topics
- Module Architecture
- Getting Started with the Demo Module
- Creating a New Java Module
- Deprecated Components
- The LabKey Server Container
- CSS Design Guidelines
- Implementing Actions and Views
- Implementing API Actions
- Integrating with the Pipeline Module
- Integrating with the Experiment Module
- GWT Integration
- GWT Remote Services
- Java Testing Tips
Module Architecture
Deploy Modules
At deployment time, a LabKey module consists of a single .module file. The .module file bundles the webapp resources (static content such as .GIF and .JPEG files, JavaScript files, SQL scripts, etc), class files (inside .jar files), and so forth.
The built .module file should be copied into your /modules directory. This directory is usually a sibling directory to the webapp directory.
At server startup time, LabKey Server extracts the modules so that it can find all the required files. It also cleans up old files that might be left from modules that have been deleted from the modules directory.
Build Modules
The build process for a module produces a .module file and copies it into the deployment directory. The standalone_build.xml file can be used for modules where the source code resides outside the standard LabKey source tree. It's important to make sure that you don't have the VM parameter -Dproject.root specified if you're developing this way or LabKey won't find all the files it loads directly from the source tree in dev mode (such as .sql and .gm files).
The create_module Ant target will prompt you for the name of a new module and a location on the file system where it should live. It then creates a minimal module that's an easy starting point for development. You can add the .IML file to your IntelliJ project and you're up and running. Use the build.xml file in the module's directory to build it.
Each module is built independently of the others. All modules can see shared classes, like those in API or third-party JARs that get copied into WEB-INF/lib. However, modules cannot see one another's classes. If two modules need to communicate with each other, they must do so through interfaces defined in the LabKey Server API, or placed in a module's own api-src directory. Currently there are many classes that are in the API that should be moved into the relevant modules. As a long-term goal, API should consist primarily of interfaces and abstract classes through which modules talk to each other. Individual modules can place third-party JARs in their lib/ directory.
Dependencies
The LabKey Server build process enforces that modules and other code follow certain dependency rules. Modules cannot depend directly on each other's implementations, and the core API cannot depend on individual modules' code. A summary of the allowed API/implementation dependencies is shown here: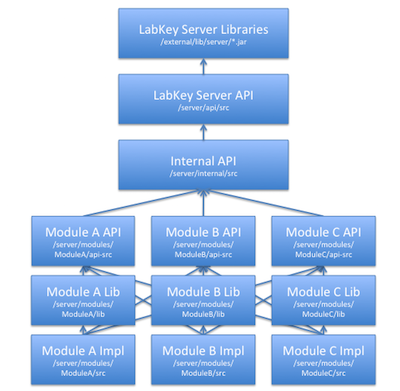
Upgrade Modules
See Upgrade Modules.
Delete Modules
To delete an unused module, delete both the .module file and the expanded directory of the same name from your deployment. The module may be in either the /modules or /externalModules directory.
Getting Started with the Demo Module
Before you get started, you need to either enlist in the version control project or download the source code. You will then need to set up your development environment to build the source code.
About the Demo Module
The Demo module is a simple sample module that displays names and ages for some number of individuals. Its purpose is to demonstrate some of the basic data display and manipulation functionality available in LabKey Server.
You can enable the Demo module in a project or folder to try it out:
- Select Admin -> Folder -> Management and choose the Folder Type tab
- Enable the Demo module
- Add the Demo Summary web part to your project or folder. A web part is an optional component that can provide a summary of the data contained in your module.
A Tour of the Demo Module
In the following sections, we'll examine the different files and classes that make up the Demo module.
Take a look at the source code at: <labkey-home>\modules. The modules\ directory contains the source code for all of the modules, each sub-directory is an individual module.
The LabKey Server web application uses a model-view-controller (MVC) architecture based on Spring.
You may also want to look at the database component of the Demo module. The Person table stores data for the Demo module.
The Object Model (Person Class)
The Person class comprises the object model for the Demo module. The Person class can be found in the org.labkey.demo.model package (and, correspondingly, in the <labkey-home>\modules\server\demo\src\org\labkey\demo\model directory). It provides methods for setting and retrieving Person data from the Person table. Note that the Person class does not retrieve or save data to the database itself, but only stores in memory data that is to be saved or has been retrieved. The Person class extends the Entity class, which contains general methods for working with objects that are stored as rows in a table in the database.
The Controller File (DemoController Class)
Modules have one or more controller classes, which handle the flow of navigation through the UI for the module. A controller class manages the logic behind rendering the HTML on a page within the module, submitting form data via both GET and POST methods, handling input errors, and navigating from one action to the next.
A Controller class is a Java class that defines individual action classes, all of which are auto-registered with the controller's ActionResolver. Action classes can also be defined outside the controller, in which case they must be registered with the ActionResolver. Action classes are annotated to declare permissions requirements.
The controller for the Demo module, DemoController.java, is located in the org.labkey.demo package (that is, in <labkey-home>\server\modules\demo\src\org\labkey\demo). If you take a look at some of the action classes in the DemoController class, you can see how the controller manages the user interface actions for the module. For example, the BeginAction in the DemoController displays data in a grid format. It doesn't write out the HTML directly, but instead calls other methods that handle that task. The InsertAction class displays a form for inserting new Person data when GET is used and calls the code that handles the database insert operation when POST is used.
A module's controller class should extend the SpringActionController class, Labkey's implementation of the Spring Controller class.
The primary controller for a module is typically named <module-name>Controller.
The Module View
The module controller renders the module user interface and also handles input from that user interface. Although you can write all of the necessary HTML from within the controller, we recommend that you separate out the user interface from the controller in most cases and use the LabKey Server rendering code to display blocks of HTML. LabKey Server primarily uses JSP files templates to render the module interface.
The bulkUpdate.jsp File
The bulkUpdate.jsp file displays an HTML form that users can use to update more than one row of the Person table at a time. BulkUpdateAction renders the bulkUpdate.jsp file and accepts posts from that HTML form. The data submitted by the user is passed to handlePost() as values on an object of type BulkUpdateForm. The form values are accessible via getters and setters on the BulkUpdateForm class that are named to correspond to the inputs on the HTML form.
The bulkUpdate.jsp file provides one example of how you can create a user interface to your data within your module. Keep in mind that you can take advantage of a lot of the basic data functionality that is already built into LabKey Server, described elsewhere in this section, to make it easier to build your module. For example, the DataRegion class provides an easy-to-use data grid with built-in sorting and filtering.
The DemoWebPart Class
The DemoWebPart class is located in the org.labkey.demo.view package. It comprises a simple web part for the demo module. This web part can be displayed only on the Portal page. It provides a summary of the data that's in the Demo module by rendering the demoWebPart.jsp file. An object of type ViewContext stores in-memory values that are also accessible to the JSP page as it is rendering.
The web part class is optional, although most modules have a corresponding web part.
The demoWebPart.jsp File
The demoWebPart.jsp file displays Person data on an HTML page. The JSP retrieves data from the ViewContext object in order to render that data in HTML.
The Data Manager Class (DemoManager Class)
The data manager class contains the logic for operations that a module performs against the database, including retrieving, inserting, updating, and deleting data. It handles persistence and caching of objects stored in the database. Although database operations can be called from the controller, as a design principle we recommend separating this layer of implementation from the navigation-handling code.
The data manager class for the Demo module, the DemoManager class, is located in the org.labkey.demo package. Note that the DemoManager class makes calls to the LabKey Server table layer, rather than making direct calls to the database itself.
The Module Class (DemoModule Class)
The DemoModule class is located in the org.labkey.demo package. It extends the DefaultModule class, which is an implementation of the Module interface. The Module interface provides generic functionality for all modules in LabKey Server and manages how the module plugs into the LabKey Server framework and how it is versioned.
The only requirement for a module is that it implement the Module interface. However, most modules have additional classes like those seen in the Demo module.
The Schema Class (DemoSchema Class)
The DemoSchema class is located in the org.labkey.demo package. It provides methods for accessing the schema of the Person table associated with the Demo module. This class abstracts schema information for this table, so that the schema can be changed in just one place in the code.
Database Scripts
The <labkey-home>\server\modules\demo\webapp\demo\scripts directory contains two subdirectories, one for PostgreSQL and one for Microsoft SQL Server. These directories contain functionally equivalent scripts for creating the Person table on the respective database server.
Note that there are a set of standard columns that all database tables in LabKey Server must include. These are:
- _ts: the timestamp column
- RowId: an autogenerated integer field that serves as the primary key
- CreatedBy: a user id
- Created: a date/time column
- ModifiedBy: a user id
- Modified: a date/time column
- Owner: a user id
Creating a New Java Module
The main build.xml file on your LabKey Server contains an Ant target called create_module. This target makes it easy to create a template Java module with the correct file structure and template Controller classes. We recommend using it instead of trying to copy an existing module, as renaming a module requires editing and renaming many files.
When you invoke the create_module target, it will prompt you for two things:
- The module name. This should be a single word (or multiple words concatenated together), for example MyModule, ProjectXAssay, etc.
- A directory in which to put the files.
- "Test"
- "C:\labkey\server\localModules\Test"
C:\labkey\server\localModules\Test
│ module.properties
│ Test.iml
│
├───lib
├───resources
│ ├───schemas
│ │ │ test.xml
│ │ │
│ │ └───dbscripts
│ │ ├───postgresql
│ │ │ test-XX.XX-YY.YY.sql
│ │ │
│ │ └───sqlserver
│ │ test-XX.XX-YY.YY.sql
│ │
│ └───web
└───src
└───org
└───labkey
└───test
│ TestContainerListener.java
│ TestController.java
│ TestManager.java
│ TestModule.java
│ TestSchema.java
│
└───view
hello.jsp
IntelliJ .iml file
If you are using IntelliJ, you can import MyModule.iml as an IntelliJ module to add your LabKey Server module to the IntelliJ project.
lib directory
JAR files required by your module but not already part of the LabKey Server distribution can be added to the ./lib directory. At compile time and run time, they will be visible to your module but not to the rest of the system. This means that different modules may use different versions of library JAR files.
Manager class
In LabKey Server, the Manager classes encapsulate much of the business logic for the module. Typical examples include fetching objects from the database, inserting, updating, and deleting objects, and so forth.
Module class
This is the entry point for LabKey Server to talk to your module. Exactly one instance of this class will be instantiated. It allows your module to register providers that other modules may use.
Schema class
Schema classes provide places to hook in to the LabKey Server Table layer, which provides easy querying of the database and object-relational mapping.
Schema XML file
This provides metadata about your database tables and views. In order to pass the developer run test (DRT), you must have entries for every table and view in your database schema. To regenerate this XML file, see Modules: Database Transition Scripts. For more information about the DRT, see Check in to the Source Project.
Controller class
This is a subclass of SpringActionController that links requests from a browser to code in your application.
web directory
All of that static web content that will be served by Tomcat should go into this directory. These items typically include things like .gif and .jpg files. The contents of this directory will be combined with the other modules' webapp content, so we recommend adding content in a subdirectory to avoid file name conflicts.
.sql files
These files are the scripts that create and update your module's database schema. They are automatically run at server startup time. See the Modules: SQL Scripts for details on how to create and modify database tables and views. LabKey Server currently supports Postgres and Microsoft SQL Server.
module.properties
At server startup time, LabKey Server uses this file to determine your module's name, class, and dependencies.
Deploy the Java Module
The main build target will build and deploy your custom module, assuming its source directory is referenced in the "standard.modules" file (either explicitly or implicitly via wildcards). The main build will compile your Java files and JSPs, package all code and resources into a .module file, and deploy it to the server.
Add a Module API
A module may define its own API which is available to the implementations of other modules. To add an API to an existing module:- Create a new api-src directory in the module's root.
- In IntelliJ, File->New Module. Choose Java as the module type. Call it MODULENAME-API, make the module's api-src directory the content root, and use the root of the module as the module file location.
- In IntelliJ, File->Project Structure. Select your new API module from the list. In the Sources tab, remove the "src" directory as a source root, and make the api-src directory as a source root. In the Dependencies tab, add a Module Dependency on the "Internal" module and check the box to Export it. Find your original module from the list. Remove the dependency on the Internal module, and add a Module Dependency on your new API module.
- Remove the "src" directory under the api-src directory.
- Create a new package under your api-src directory, "org.labkey.MODULENAME.api" or similar.
- Add Java classes to the new package, and reference them from within your module.
- Add a Module Dependency to any other modules that depend on your module's API.
- Develop and test.
- Commit your new Java source files, the new .IML file, any .IML files for existing modules that you changed, and the reference to your new .IML API in LabKey.ipr.
The LabKey Server Container
The Container on the URL
The container hierarchy is always included in the URL, following the name of the controller. For example, the URL below shows that it is in the /Documentation folder beneath the /home project:
https://www.labkey.org/home/Documentation/wiki-page.view?name=buildingModule
The getExtraPath() method of the ViewURLHelper class returns the container path from the URL. On the Container object, the getPath() method returns the container's path.
The Root Container
LabKey Server also has a root container which is not apparent in the user interface, but which contains all other containers. When you are debugging LabKey Server code, you may see the Container object for the root container; its name appears as "/".
In the core.Containers table in the LabKey Server database, the root container has a null value for both the Parent and the Name field.
You can use the isRoot() method to determine whether a given container is the root container.
Projects Versus Folders
Given that they are both objects of type Container, projects and folders are essentially the same at the level of the implementation. A project will always have the root container as its parent, while a folder's parent will be either a project or another folder.
You can use the isProject() method to determine whether a given container is a project or a folder.
Useful Classes and Methods
Container Class Methods
The Container class represents a given container and persists all of the properties of that container. Some of the useful methods on the Container class include:
- getName(): Returns the container name
- getPath(): Returns the container path
- getId(): Returns the GUID that identifies this container
- getParent(): Returns the container's parent container
- hasPermission(user, perm): Returns a boolean indicating whether the specified user has the given level of permissions on the container
The ContainerManager class includes a number of static methods for managing containers. Some useful methods include:
- create(container, string): Creates a new container
- delete(container): Deletes an existing container
- ensureContainer(string): Checks to make sure the specified container exists, and creates it if it doesn't
- getForId(): Returns the container with this EntityId (a GUID value)
- getForPath(): Returns the container with this path
The controller class in your LabKey Server module extends the ViewController class, which provides the getContainer() method. You can use this method to retrieve the Container object corresponding to the container in which the user is currently working.
Implementing Actions and Views
Actions are the "surface area" of the server: everything you invoke on the server, whether a view on data or a manipulation of data, is some action or set of actions. An Action is implemented using the Model-View-Controller paradigm, where:
- the Model is implemented as one or more Java classes, such as standard JavaBean classes
- the View is implemented as JSPs, or other technologies
- the Controller is implemented as Java action classes
Views are typically implemented in parent-child relationships, such that a page is built from a template view that wraps one or more body views. Views often render other views, for example, one view per pane or a series of similar child views. Views are implemented using a variety of different rendering technologies; if you look at the subclasses of HttpView and browse the existing controllers you will see that views can be written using JSP, GWT, out.print() from Java code, etc. (Note that most LabKey developers write JSPs to create new views. The JSP syntax is familiar and supported by all popular IDEs, JSPs perform well, and type checking & compilation increase reliability.)
Action Life Cycle
What happens when you submit to an Action in LabKey Server? The typical life cycle looks like this:
- ViewServlet receives the request and directs it to the appropriate module.
- The module passes the request to the appropriate Controller which then invokes the requested action.
- The action verifies that the user has permission to invoke it in the current folder. (If the user is not assigned an appropriate role in the folder then the action will not be invoked.) Action developers typically declare required permissions via a @RequiresPermission() annotation.
- The Spring framework instantiates the Form bean associated with the action and "binds" parameter values to it. In other words, it matches URL parameters names to bean property names; for each match, it converts the parameter value to the target data type, performs basic validation, and sets the property on the form by calling the setter.
- The Controller now has data, typed and validated, that it can work with. It performs the action, and typically redirects to a results page, confirmation page, or back to the same page.
Example: Hello World JSP View
The following action takes a user to a static "Hello World" JSP view.
helloWorld.jsp:
<%= h("Hello, World!") %>HelloWorldAction:
// If the user does not have Read permissions, the action will not be invoked.
@RequiresPermission(ReadPermission.class)
public class HelloWorldAction extends SimpleViewAction
{
@Override
public ModelAndView getView(Object o, BindException errors) throws Exception
{
JspView view = new JspView("/org/labkey/javatutorial/view/helloWorld.jsp");
view.setTitle("Hello World");
return view;
}
@Override
public NavTree appendNavTrail(NavTree root)
{
return root;
}
}
The HelloWorld Action is called with this URL:
Example: Submitting Forms to an Action
The following action processes a form submitted by the user.
helloSomeone.jsp
This JSP is for submitting posts, and displaying responses, on the same page:
<%@ taglib prefix="labkey" uri="http://www.labkey.org/taglib" %>
<%@ page import="org.labkey.api.view.HttpView"%>
<%@ page import="org.labkey.javatutorial.JavaTutorialController" %>
<%@ page import="org.labkey.javatutorial.HelloSomeoneForm" %>
<%@ page extends="org.labkey.api.jsp.JspBase" %>
<%
HelloSomeoneForm form = (HelloSomeoneForm) HttpView.currentModel();
%>
<labkey:errors />
<labkey:form method="POST" action="<%=urlFor(JavaTutorialController.HelloSomeoneAction.class)%>">
<h2>Hello, <%=h(form.getName()) %>!</h2>
<table width="100%">
<tr>
<td class="labkey-form-label">Who do you want to say 'Hello' to next?: </td>
<td><input name="name" value="<%=h(form.getName())%>"></td>
</tr>
<tr>
<td><labkey:button text="Go" /></td>
</tr>
</table>
</labkey:form>
Action for handling posts:
// If the user does not have Read permissions, the action will not be invoked.
@RequiresPermission(ReadPermission.class)
public class HelloSomeoneAction extends FormViewAction<HelloSomeoneForm>
{
public void validateCommand(HelloSomeoneForm form, Errors errors)
{
// Do some error handling here
}
public ModelAndView getView(HelloSomeoneForm form, boolean reshow, BindException errors) throws Exception
{
return new JspView<>("/org/labkey/javatutorial/view/helloSomeone.jsp", form, errors);
}
public boolean handlePost(HelloSomeoneForm form, BindException errors) throws Exception
{
return true;
}
public ActionURL getSuccessURL(HelloSomeoneForm form)
{
// Redirect back to the same action, adding the submitted value to the URL.
ActionURL url = new ActionURL(HelloSomeoneAction.class, getContainer());
url.addParameter("name", form.getName());
return url;
}
public NavTree appendNavTrail(NavTree root)
{
root.addChild("Say Hello To Someone");
return root;
}
}
Below is the form used to convey the URL parameter value to the Action class. Note that the form follows a standard JavaBean format. The Spring framework attempts to match URL parameter names to property names in the form. If it finds matches, it interprets the URL parameters according to the data types it finds in the Bean property and performs basic data validation on the values provided on the URL:
package org.labkey.javatutorial;
public class HelloSomeoneForm
{
public String _name = "World";
public void setName(String name)
{
_name = name;
}
public String getName()
{
return _name;
}
}
URL that invokes the action in the home project:
Example: Export as Script Action
This action exports a query as a re-usable script, either as JavaScript, R, Perl, or SAS. (The action is surfaced in the user interface on a data grid, at Export > Script.)
public static class ExportScriptForm extends QueryForm
{
private String _type;
public String getScriptType()
{
return _type;
}
public void setScriptType(String type)
{
_type = type;
}
}
@RequiresPermission(ReadPermission.class)
public class ExportScriptAction extends SimpleViewAction<ExportScriptForm>
{
public ModelAndView getView(ExportScriptForm form, BindException errors) throws Exception
{
ensureQueryExists(form);
return ExportScriptModel.getExportScriptView(QueryView.create(form, errors),
form.getScriptType(), getPageConfig(), getViewContext().getResponse());
}
public NavTree appendNavTrail(NavTree root)
{
return null;
}
}
Example: Delete Cohort
The following action deletes a cohort category from a study (provided it is an empty cohort). It then redirects the user back to the Manage Cohorts page.
@RequiresPermission(AdminPermission.class)
public class DeleteCohortAction extends SimpleRedirectAction<CohortIdForm>
{
public ActionURL getRedirectURL(CohortIdForm form) throws Exception
{
CohortImpl cohort = StudyManager.getInstance().getCohortForRowId(getContainer(), getUser(), form.getRowId());
if (cohort != null && !cohort.isInUse())
StudyManager.getInstance().deleteCohort(cohort);
return new ActionURL(CohortController.ManageCohortsAction.class, getContainer());
}
}
Packaging JSPs
JSPs can be placed anywhere in the src directory, but by convention they are often placed in the view directory, as shown below:
mymodule
├───lib
├───resources
└───src
└───org
└───labkey
└───javatutorial
│ HelloSomeoneForm.java
│ JavaTutorialController.java
│ JavaTutorialModule.java
└───view
helloSomeone.jsp
helloWorld.jsp
Implementing API Actions
Overview
This page describes how to implement API actions within the LabKey Server controller classes. It is intended for Java developers building their own modules or working within the LabKey Server source code.
API actions build upon LabKey’s controller/action design. They include the “API” action base class whose derived action classes interact with the database or server functionality. These derived actions return raw data to the base classes, which serialize raw data into one of LabKey’s supported formats.
Leveraging the current controller/action architecture provides a range of benefits, particularly:
- Enforcement of user login for actions that require login, thanks to reuse of LabKey’s existing, declarative security model (@RequiresPermission annotations).
- Reuse of many controllers’ existing action forms, thanks to reuse of LabKey’s existing Spring-based functionality for binding request parameters to form beans.
API Action Design Rules
In principle, actions are autonomous, may be named, and can do, whatever the controller author wishes. However, in practice, we suggest adhering to the following general design rules when implementing actions:
- Action names should be named with a verb/noun pair that describes what the action does in a clear and intuitive way (e.g., getQuery, updateList, translateWiki, etc.).
- Insert, update, and delete of a resource should all be separate actions with appropriate names (e.g., getQuery, updateRows, insertRows, deleteRows), rather than a single action with a parameter to indicate the command.
- Wherever possible, actions should remain agnostic about the request and response formats. This is accomplished automatically through the base classes, but actions should refrain from reading the post body directly or writing directly to the HttpServletResponse unless they absolutely need to.
- For security reasons, ApiActions that respond to GET should not mutate the database or otherwise change server state. ApiActions that change state (e.g., insert, update, or delete actions) should respond to POST and extend MutatingApiAction.
API Actions
An APIAction is a Spring-based action that derives from the abstract base class org.labkey.api.action.ApiAction. API actions do not implement the getView() or appendNavTrail() methods that view actions do. Rather, they implement the execute method. MyForm is a simple bean intended to represent the parameters sent to this action.
@RequiresPermission(ReadPermission.class)
public class GetSomethingAction extends ApiAction<MyForm>
{
public ApiResponse execute(MyForm form, BindException errors) throws Exception
{
ApiSimpleResponse response = new ApiSimpleResponse();
// Get the resource...
// Add it to the response...
return response;
}
}
JSON Example
A basic API action class looks like this:
@RequiresPermission(ReadPermission.class)
public class ExampleJsonAction extends ApiAction<Object>
{
public ApiResponse execute(Object form, BindException errors) throws Exception
{
ApiSimpleResponse response = new ApiSimpleResponse();
response.put("param1", "value1");
response.put("success", true);
return response;
}
}
A URL like the following invokes the action:
Returning the following JSON object:
{
"success" : true,
"param1" : "value1"
}Example: Set Display for Table of Contents
@RequiresLogin
public class SetTocPreferenceAction extends MutatingApiAction<SetTocPreferenceForm>
{
public static final String PROP_TOC_DISPLAYED = "displayToc";
public ApiResponse execute(SetTocPreferenceForm form, BindException errors)
{
//use the same category as editor preference to save on storage
PropertyManager.PropertyMap properties = PropertyManager.getWritableProperties(
getUser(), getContainer(),
SetEditorPreferenceAction.CAT_EDITOR_PREFERENCE, true);
properties.put(PROP_TOC_DISPLAYED, String.valueOf(form.isDisplayed()));
PropertyManager.saveProperties(properties);
return new ApiSimpleResponse("success", true);
}
}
Execute Method
public ApiResponse execute(FORM form, BindException errors) throws Exception
In the execute method, the action does whatever work it needs to do and responds by returning an object that implements the ApiResponse interface. This ApiResponse interface allows actions to respond in a format-neutral manner. It has one method, getProperties(), that returns a Map<String,Object>. Two implementations of this interface are available: ApiSimpleResponse, which should be used for simple cases; and ApiQueryResponse, which should be used for returning the results of a QueryView.
ApiSimpleResponse has a number of constructors that make it relatively easy to send back simple response data to the client. For example, to return a simple property of “rowsUpdated=5”, your return statement would look like this:
return new ApiSimpleResponse("rowsUpdated", rowsUpdated);
where rowsUpdated is an integer variable containing the number of rows updated. Since ApiSimpleResponse derives from HashMap<String, Object>, you may put as many properties in the response as you wish. A property value may also be a nested Map, Collection, or array.
The ApiAction base class takes care of serializing the response in the JSON appropriate format.
Although nearly all API actions return an ApiResponse object, some actions necessarily need to return data in a specific format, or even binary data. In these cases, the action can use the HttpServletResponse object directly, which is available through getViewContext().getReponse(), and simply return null from the execute method.
Form Parameter Binding
If the request uses a standard query string with a GET method, form parameter binding uses the same code as used for all other view requests. However, if the client uses the POST method, the binding logic depends on the content-type HTTP header. If the header contains the JSON content-type (“application/json”), the ApiAction base class parses the post body as JSON and attempts to bind the resulting objects to the action’s form. This code supports nested and indexed objects via the BeanUtils methods.
For example, if the client posts JSON like this:
{ "name": "Lister",
"address": {
"street": "Top Bunk",
"city": “Red Dwarf",
"state": “Deep Space"},
"categories” : ["unwashed", "space", "bum"]
}The form binding uses BeanUtils to effectively make the following calls via reflection:
form.setName("Lister");
form.getAddress().setStreet("Top Bunk");
form.getAddress().setCity("Red Dwarf");
form.getAddress().setState("Deep Space");
form.getCategories().set(0) = "unwashed";
form.getCategories().set(1) = "space";
form.getCategories().set(2) = "bum";
Where an action must deal with the posted data in a dynamic way (e.g., the insert, update, and delete query actions), the action’s form may implement the ApiJsonForm interface to receive the parsed JSON data directly. If the form implements this interface, the binding code simply calls the setJsonObject() method, passing the parsed JSONObject instance, and will not perform any other form binding. The action is then free to use the parsed JSON data as necessary.
Jackson Marshalling (Experimental)
Experimental Feature: Instead of manually unpacking the JSONObject from .getJsonObject() or creating a response JSONObject, you may use Jackson to marshall a Java POJO form and return value. To enable Jackson marshalling, add the @Marshal(Marshaller.Jackson) annotation to your Controller or ApiAction class. When adding the @Marshal annotation to a controller, all ApiActions defined in the Controller class will use Jackson marshalling. For example,
@Marshal(Marshaller.Jackson)
@RequiresLogin
public class ExampleJsonAction extends ApiAction<MyStuffForm>
{
public ApiResponse execute(MyStuffForm form, BindException errors) throws Exception
{
// retrieve resource from the database
MyStuff stuff = ...;
// instead of creating an ApiResponse or JSONObject, return the POJO
return stuff;
}
}
Error and Exception Handling
If an API action adds errors to the errors collection or throws an exception, the base ApiAction will return a response with status code 400 and a json body using the format below. Clients may then choose to display the exception message or react in any way they see fit. For example, if an error is added to the errors collection for the "fieldName" field of the ApiAction's form class with message "readable message", the response will be serialized as:
{
"success": false,
"exception": "readable message",
"errors": [ {
"id" : "fieldName",
"msg" : "readable message",
} ]
}Integrating with the Pipeline Module
Other modules can register themselves as providing pipeline functionality, and the Pipeline module will let them indicate the types of analysis that can be done on files, as well as delegate to them to do the actual work.
Integration points
org.labkey.api.pipeline.PipelineProvider
PipelineProviders let modules hook into the Pipeline module's user interface for browsing through the file system to find files on which to operate. This is always done within the context of a pipeline root for the current folder. The Pipeline module calls updateFileProperties() on all the PipelineProviders to determine what actions should be available. Each module provides its own URL which can collect additional information from the user before kicking off any work that needs to be done.
For example, the org.labkey.api.exp.ExperimentPipelineProvider registered by the Experiment module provides actions associated with .xar and .xar.xml files. It also provides a URL that the Pipeline module associates with the actions. If the users clicks to load a XAR, the user's browser will go to the Experiment module's URL.
PipelineProviders are registered by calling org.labkey.api.pipeline.PipelineServer.registerPipelineProvider().
org.labkey.api.pipeline.PipelineJob
PipelineJobs allow modules to do work relating to a particular piece of analysis. PipelineJobs sit in a queue until the Pipeline module determines that it is their turn to run. The Pipeline module then calls the
PipelineJob's run() method. The PipelineJob base class provides logging and status functionality so that implementations can inform the user of their progress.
The Pipeline module attempts to serialize the PipelineJob object when it is submitted to the queue. If the server is restarted while there are jobs in the queue, the Pipeline module will look for all the jobs that were not in the COMPLETE or ERROR state, deserialize the PipelineJob objects from disk, and resubmit them to the queue. A PipelineJob implementation is responsible for restarting correctly if it is interrupted in the middle of processing. This might involve resuming analysis at the point it was interrupted, or deleting a partially loaded file from the database before starting to load it again.
For example, the org.labkey.api.exp.ExperimentPipelineJob provided by the Experiment module knows how to parse and load a XAR file. If the input file is not a valid XAR, it will put the job into an error state and write the reason to the log file.
PipelineJobs do not need to be explicitly registered with the Pipeline module. Other modules can add jobs to the queue using the org.labkey.api.pipeline.PipelineService.queueJob() method.
Integrating with the Experiment Module
Integration points
org.labkey.api.exp.ExperimentDataHandler
The ExperimentDataHandler interface allows a module to handle specific kinds of files that might be present in a XAR. When loading from a XAR, the Experiment module will keep track of all the data files that it encounters. After the general, Experiment-level information is fully imported, it will call into the ExperimentDataHandlers that other modules have registered. This gives other modules a chance to load data into the database or otherwise prepare it for later display. The XAR load will fail if an ExperimentDataHandler throws an ExperimentException, indicating that the data file was not as expected.
Similarly, when exporting a set of runs as a XAR, the Experiment module will call any registered ExperimentDataHandlers to allow them to transform the contents of the file before it is written to the compressed archive. The default exportFile() implementation, provided by AbstractExperimentDataHandler, simply exports the file as it exists on disk.
The ExperimentDataHandlers are also interrogated to determine if any modules provide UI for viewing the contents of the data files. By default, users can download the content of the file, but if the ExperimentDataHandler provides a URL, it will also be available. For example, the MS2 module provides an ExperimentDataHandler that hands out the URL to view the peptides and proteins for a .pep.xml file.
Prior to deleting a data object, the Experiment module will call the associated ExperimentDataHandler so that it can do whatever cleanup is necessary, like deleting any rows that have been inserted into the database for that data object.
ExperimentDataHandlers are registered by implementing the getDataHandlers() method on Module.
org.labkey.api.exp.RunExpansionHandler
RunExpansionHandlers allow other modules to modify the XML document that describes the XAR before it is imported. This means that modules have a chance to run Java code to make decisions on things like the number and type of outputs for a ProtocolApplication based on any criteria they desire. This provides flexibility beyond just what is supported in the XAR schema for describing runs. They are passed an XMLBeans representation of the XAR.
RunExpansionHandlers are registered by implementing the getRunExpansionHandlers() method on Module.
org.labkey.api.exp.ExperimentRunFilter
ExperimentRunFilters let other modules drive what columns are available when viewing particular kinds of runs in the experiment run grids in the web interface. The filter narrows the list of runs based on the runs' protocol LSID.
Using the Query module, the ExperimentRunFilter can join in additional columns from other tables that may be related to the run. For example, for MS2 search runs, there is a row in the MS2Runs table that corresponds to a row in the exp.ExperimentRun table. The MS2 module provides ExperimentRunFilters that tell the Experiment module to use a particular virtual table, defined in the MS2 module, to display the MS2 search runs. This virtual table lets the user select columns for the type of mass spectrometer used, the name of the search engine, the type of quantitation run, and so forth. The virtual tables defined in the MS2 schema also specify the set of columns that should be visible by default, meaning that the user will automatically see some of files that were the inputs to the run, like the FASTA file and the mzXML file.
ExperimentRunFilters are registered by implementing the getExperimentRunFilters() method on Module.
Generating and Loading XARs
When a module does data analysis, typically performed in the context of a PipelineJob, it should generally describe the work that it has done in a XAR and then cause the Experiment module to load the XAR after the analysis is complete.
It can do this by creating a new ExperimentPipelineJob and inserting it into the queue, or by calling org.labkey.api.exp.ExperimentPipelineJob.loadExperiment(). The module will later get callbacks if it has registered the appropriate ExperimentDataHandlers or RunExpansionHandlers.
API for Creating Simple Protocols and Experiment Runs
Version 2.2 of LabKey Server introduces an API for creating simple protocols and simple experiment runs that use those protocols. It is appropriate for runs that start with one or more data/material objects and output one or more data/material objects after performing a single logical step.
To create a simple protocol, call org.labkey.api.exp.ExperimentService.get().insertSimpleProtocol(). You must pass it a Protocol object that has already been configured with the appropriate properties. For example, set its description, name, container, and the number of input materials and data objects. The call will create the surrounding Protocols, ProtocolActions, and so forth, that are required for a full fledged Protocol.
To create a simple experiment run, call org.labkey.api.exp.ExperimentService.get().insertSimpleExperimentRun(). As with creating a simple Protocol, you must populate an ExperimentRun object with the relevant properties. The run must use a Protocol that was created with the insertSimpleProtocol() method. The run must have at least one input and one output. The call will create the ProtocolApplications, DataInputs, MaterialInputs, and so forth that are required for a full-fledged ExperimentRun.
Using SQL in Java Modules
Ways to Work with SQL
Options for working with SQL from Java code:
Table Class
Using Table.insert()/update()/delete() with a simple Java class/bean works well when you want other code to be able to work with the class, and the class fields map directly with what you're using in the database. This approach usually results in the least lines of code to accomplish the goal. See the demoModule for an example of this approach.SQLFragment/SQLExecutor
SQLFragment/SQLExecutor is a good approach when you need more control over the SQL you're generating. It's also used for operations that work on multiple rows at a time.Prepared SQL Statements
Use prepared statements when you're dealing with many data rows and want the performance gain from being able to reuse the same statement with different values.Client-Side Options
You can also develop SQL applications without needing any server-side Java code by using the LABKEY.Query.saveRows() and related APIs from JavaScript code in the client. In this scenario, you'd expose your table as part of a schema, and rely on the default server implementation. This approach gives you the least control over the SQL that's actually used.Utility Functions
LabKey Server provides a number of SQL function extensions to help Java module developers:
- access various properties
- keep Java code and SQL queries in sync
moduleProperty(MODULE_NAME, PROPERTY_NAME)
Returns a module property, based on the module and property names. Arguments are strings, so use single quotes not double.
Examples
moduleProperty('EHR','EHRStudyContainer')You can use the virtual "Site" schema to specify a full container path, such as '/home/someSubfolder' or '/Shared':
SELECT *
FROM Site.{substitutePath moduleProperty('EHR','EHRStudyContainer')}.study.myQuery
javaConstant(FULLY_QUALIFIED_CLASS_AND_FIELD_NAME)
Provides access to public static final variable values. The argument value should be a string.
Fields must be either be on classes in the java.lang package, or tagged with the org.labkey.api.query.Queryable annotation to indicate they allow access through this mechanism. Other fields types are not supported.
Examples
javaConstant('java.lang.Integer.MAX_VALUE')
javaConstant('org.labkey.mymodule.MyConstants.MYFIELD')To allow access to MYFIELD, tag the field with the annotation @Queryable:
public class MyConstants
{
@Queryable
public static final String MYFIELD = "some value";
}
GWT Integration
LabKey Server uses the Google Web Toolkit (GWT) to create web pages with rich UI. GWT compiles java code into JavaScript that runs in a browser. For more information about GWT see the GWT home page.
We have done some work to integrate GWT into the LabKey framework:
- The org.labkey.api.gwt.Internal GWT module can be inherited by all other GWT modules to include tools that allow GWT clients to connect back to the LabKey server more easily.
- There is a special incantation to integrate GWT into a web page. The org.labkey.api.view.GWTView class allows a GWT module to be incorporated in a standard LabKey web page.
- GWTView also allows passing parameters to the GWT page. The org.labkey.api.gwt.client.PropertyUtil class can be used by the client to retrieve these properties.
- GWT supports asynchronous calls from the client to servlets. To enforce security and the module architecture a few classes have been provided to allow these calls to go through the standard LabKey security and PageFlow mechanisms.
- The client side org.labkey.api.gwt.client.ServiceUtil class enables client->server calls to go through a standard LabKey action implementation.
- The server side org.labkey.api.gwt.server.BaseRemoteService class implements the servlet API but can be configured with a standard ViewContext for passing a standard LabKey url and security context.
- Create an action in your controller that instantiates your servlet (which should extend BaseRemoteService) and calls doPost(getRequest(), getResponse()). In most cases you can simply create a subclass of org.labkey.api.action.GWTServiceAction and implement the createService() method.
- Use ServiceUtil.configureEndpoint(service, "actionName") to configure client async service requests to go through your PageFlow action on the server.
Examples of this can be seen in the study.designer and plate.designer packages within the Study module.
The checked-in jars allow GWT modules within Labkey modules to be built automatically. Client-side classes (which can also be used on the server) are placed in a gwtsrc directory parallel to the standard src directory in the module.
While GWT source can be built automatically, effectively debugging GWT modules requires installation of the full GWT toolkit (we are using 2.5.1 currently). After installing the toolkit you can debug a page by launching GWT's custom client using the class com.google.gwt.dev.DevMode, which runs java code rather than the cross-compiled javascript. The debug configuration is a standard java app with the following requirements
- gwt-user.jar and gwt-dev.jar from your full install need to be on the runtime classpath. (Note: since we did not check in client .dll/.so files, you need to point a manually installed local copy of the GWT development kit.)
- the source root for your GWT code needs to be on the runtime classpath
- the source root for the LabKey GWT internal module needs to be on the classpath
- Main class is com.google.gwt.dev.DevMode
- Program parameters should be something like this:
-noserver -startupUrl "http://localhost:8080/labkey/query/home/metadataQuery.view?schemaName=issues&query.queryName=Issues" org.labkey.query.metadata.MetadataEditor
- -noserver tells the GWT client not to launch its own private version of tomcat
- the URL is the url you would like the GWT client to open
- the last parameter is the module name you want to debug
For example, here is a configuration from a developer's machine. It assumes that the LabKey Server source has is at c:\labkey and that the GWT development kit has been extracted to c:\JavaAPIs\gwt-windows-2.5.1. It will work with GWT code from the MS2, Experiment, Query, List, and Study modules.
- Main class: com.google.gwt.dev.DevMode
- VM parameters:
-classpath C:/labkey/server/internal/gwtsrc;C:/labkey/server/modules/query/gwtsrc;C:/labkey/server/modules/study/gwtsrc;C:/labkey/server/modules/ms2/gwtsrc;C:/labkey/server/modules/experiment/gwtsrc;C:/JavaAPIs/gwt-2.5.1/gwt-dev.jar; C:/JavaAPIs/gwt-2.5.1/gwt-user.jar;c:\labkey\external\lib\build\gxt.jar;C:/labkey/server/modules/list/gwtsrc;C:\labkey\external\lib\server\gwt-dnd-3.2.0.jar
- Program parameters:
-noserver -startupUrl "http://localhost:8080/labkey/query/home/metadataQuery.view?schemaName=issues&query.queryName=Issues" org.labkey.query.metadata.MetadataEditor
- Working directory: C:\labkey\server
- Use classpath and JDK of module: QueryGWT
A note about upgrading to future versions of GWT: As of GWT 2.6.0 (as of this writing, the current release), GWT supports Java 7 syntax. It also stops building permutations for IE 6 and 7 by default. However, it introduces a few breaking API changes. This means that we would need to move to GXT 3.x, which is unfortunately a major upgrade and requires significant changes to our UI code that uses it.
GWT Remote Services
Integrating GWT Remote services is a bit tricky within the LabKey framework. Here's a technique that works.
1. Create a synchronous service interface in your GWT client code:
import com.google.gwt.user.client.rpc.RemoteService;
import com.google.gwt.user.client.rpc.SerializableException;
public interface MyService extends RemoteService
{
String getSpecialString(String inputParam) throws SerializableException;
}
2. Create the asynchronous counterpart to your synchronous service interface. This is also in client code:
import com.google.gwt.user.client.rpc.AsyncCallback;
public interface MyServiceAsync
{
void getSpecialString(String inputParam, AsyncCallback async);
}
3. Implement your service within your server code:
import org.labkey.api.gwt.server.BaseRemoteService;
import org.labkey.api.gwt.client.util.ExceptionUtil;
import org.labkey.api.view.ViewContext;
import com.google.gwt.user.client.rpc.SerializableException;
public class MyServiceImpl extends BaseRemoteService implements MyService
{
public MyServiceImpl(ViewContext context)
{
super(context);
}
public String getSpecialString(String inputParameter) throws SerializableException
{
if (inputParameter == null)
throw ExceptionUtil.convertToSerializable(new
IllegalArgumentException("inputParameter may not be null"));
return "Your special string was: " + inputParameter;
}
}
4. Within the server Spring controller that contains the GWT action, provide a service entry point:
import org.labkey.api.gwt.server.BaseRemoteService;
import org.labkey.api.action.GWTServiceAction;
@RequiresPermission(ACL.PERM_READ)
public class MyServiceAction extends GWTServiceAction
{
protected BaseRemoteService createService()
{
return new MyServiceImpl(getViewContext());
}
}
5. Within your GWT client code, retrive the service with a method like this. Note that caching the service instance is important, since construction and configuration is expensive.
import com.google.gwt.core.client.GWT;
import org.labkey.api.gwt.client.util.ServiceUtil;
private MyServiceAsync _myService;
private MyServiceAsync getService()
{
if (_testService == null)
{
_testService = (MyServiceAsync) GWT.create(MyService.class);
ServiceUtil.configureEndpoint(_testService, "myService");
}
return _testService;
}
6. Finally, call your service from within your client code:
public void myClientMethod()
{
getService().getSpecialString("this is my input string", new AsyncCallback()
{
public void onFailure(Throwable throwable)
{
// handle failure here
}
public void onSuccess(Object object)
{
String returnValue = (String) object;
// returnValue now contains the string returned from the server.
}
});
}
Java Testing Tips
HotSwapping Java classes
Limitations
You cannot change the "shape" of a class. This means you can't add or remove member variables, methods, change the superclass, etc. This restriction may be relaxed by newer VMs someday. The VM will tell you if it can't handle the request.
You cannot change a class that hasn't been loaded by the VM already. The VM will ignore the request.
The webapp will always start up with the version of the class that was produced by the Ant build, even if you HotSwapped during an earlier debug session.
Changes to your class will be reflected AFTER the current stack has exited your method.
Workflow
These steps are the sequence in IntelliJ. Other IDEs should very similar.
- Do an Ant build.
- In IntelliJ, do Build->Make Project. This gets IntelliJ's build system primed.
- Start up Tomcat, and use the webapp so that the class you want to change is loaded (the line breakpoint icon will show a check in the left hand column once it's been loaded).
- Edit the class.
- In IntelliJ, do Build->Compile <MyClass>.java.
- If you get a dialog, tell the IDE to HotSwap and always do that in the future.
- Make your code run again. Marvel at how fast it was.
Deprecated Components
Deprecated Components
Older versions of LabKey supported components that have been deprecated. Developers creating new modules or updating existing modules should remove dependencies on these deprecated components:
- PostgreSQL 8.1, 8.2
- Microsoft SQL Server 2000, 2005, 2008 (pre-R2)
- Beehive PageFlows (ViewController, @Jpf.Action, @Jpf.Controller)
- Struts (FormData, FormFile, StrutsAttachmentFile)
- Groovy (.gm files, GroovyView, GroovyExpression, BooleanExpression)
- ACL-based permissions
Modules: Folder Types
Advanced users can define custom folder types in an XML format for easy reuse. This document explains how to define a custom folder type in your LabKey Server module. A folder type can be thought of as a template for the layout of the folder. The folder type specifies the tabs, webparts and active modules that are initially enabled in that folder.
Each folder type can provide the following:
- The name of the folder type.
- Description of the folder type.
- A list of tabs (provide a single tab for a non-tabbed folder).
- A list of the modules enabled by default for this folder.
- Whether the menu bar is enabled by default. If this is true, when the folderType is activated in a project (but not a subfolder), the menu bar will be enabled.
- The name and caption for the tab.
- An ordered list of 'required webparts'. These webparts cannot be removed.
- An ordered list of 'preferred webparts'. The webparts can be removed.
- A list of permissions required for this tab to be visible (ie. READ, INSERT, UPDATE, DELETE, ADMIN)
- A list of selectors. These selectors are used to test whether this tab should be highlighted as the active tab or not. Selectors are described in greater detail below.
Define a Custom Folder Type
Module Location
The easiest way to define a custom folder type is via a module, which is just a directory containing various kinds of resource files. Modules can be placed in the standard modules/ directory, or in the externalModules/ directory. By default, the externalModules/ directory is a peer to the modules/ directory.
To tell LabKey Server to look for external modules in a different directory, simply add the following to your VM parameters:
-Dlabkey.externalModulesDir="C:/externalModules"This will cause the server to look in C:/externalModules for module files in addition to the normal modules/ directory under the web application.
Create a directory structure like the following, replacing 'MyModule' with the name of your module. Within the folderTypes directory, any number of XML files defining new folder types can be provided.
MyModule
└───resources
└───folderTypes
Definition file name and location
Custom folder types are defined via XML files in the folderTypes directory. Folder type definition files can have any name, but must end with a ".foldertype.xml" extension. For example, the following file structure is valid:
MyModule
└───resources
└───folderTypes
myType1.foldertype.xml
myType2.foldertype.xml
myType3.foldertype.xml
Example #1
The full XML schema (XSD) for folder type XML is documented and available for download. However, the complexity of XML schema files means it is often simpler to start from an example. The following XML defines a simple folder type:
<folderType xmlns="http://labkey.org/data/xml/folderType">
<name>My XML-defined Folder Type</name>
<description>A demonstration of defining a folder type in an XML file</description>
<requiredWebParts>
<webPart>
<name>Query</name>
<location>body</location>
<property name="title" value="A customized web part" />
<property name="schemaName" value="study" />
<property name="queryName" value="SpecimenDetail" />
</webPart>
<webPart>
<name>Data Pipeline</name>
<location>body</location>
</webPart>
<webPart>
<name>Experiment Runs</name>
<location>body</location>
</webPart>
</requiredWebParts>
<preferredWebParts>
<webPart>
<name>Sample Sets</name>
<location>body</location>
</webPart>
<webPart>
<name>Run Groups</name>
<location>right</location>
</webPart>
</preferredWebParts>
<modules>
<moduleName>Experiment</moduleName>
<moduleName>Pipeline</moduleName>
</modules>
<defaultModule>Experiment</defaultModule>
</folderType>
Valid webpart names
Each <webPart> element must contain a <name> element. The example above specified that a query webpart is required via the following XML:
<requiredWebParts>
<webPart>
<name>Query</name>
Valid module names
The modules and defaultModules sections define which modules are active in the custom folder type. From the example above:
<modules>
<moduleName>Experiment</moduleName>
<moduleName>Pipeline</moduleName>
</modules>
<defaultModule>Experiment</defaultModule>
Valid module names can be found by navigating through the administrative user interface to create a new LabKey Server folder, or by selecting 'customize folder' for any existing folder. The 'customize folder' user interface includes a list of valid module names on the right-hand side.
Example #2 - Tabs
This is another example of an XML file defining a folder type:
<folderType xmlns="http://labkey.org/data/xml/folderType" xmlns:mp="http://labkey.org/moduleProperties/xml/">
<name>Laboratory Folder</name>
<description>The default folder layout for basic lab management</description>
<folderTabs>
<folderTab>
<name>overview</name>
<caption>Overview</caption>
<selectors>
</selectors>
<requiredWebParts>
</requiredWebParts>
<preferredWebParts>
<webPart>
<name>Laboratory Home</name>
<location>body</location>
</webPart>
<webPart>
<name>Lab Tools</name>
<location>right</location>
</webPart>
</preferredWebParts>
</folderTab>
<folderTab>
<name>workbooks</name>
<caption>Workbooks</caption>
<selectors>
</selectors>
<requiredWebParts>
</requiredWebParts>
<preferredWebParts>
<webPart>
<name>Workbooks</name>
<location>body</location>
</webPart>
<webPart>
<name>Lab Tools</name>
<location>right</location>
</webPart>
</preferredWebParts>
</folderTab>
<folderTab>
<name>data</name>
<caption>Data</caption>
<selectors>
<selector>
<controller>assay</controller>
</selector>
<selector>
<view>importData</view>
</selector>
<selector>
<view>executeQuery</view>
</selector>
</selectors>
<requiredWebParts>
</requiredWebParts>
<preferredWebParts>
<webPart>
<name>Data Views</name>
<location>body</location>
</webPart>
<webPart>
<name>Lab Tools</name>
<location>right</location>
</webPart>
</preferredWebParts>
</folderTab>
<folderTab>
<name>settings</name>
<caption>Settings</caption>
<selectors>
<selector>
<view>labSettings</view>
</selector>
</selectors>
<requiredWebParts>
</requiredWebParts>
<preferredWebParts>
<webPart>
<name>Lab Settings</name>
<location>body</location>
</webPart>
<webPart>
<name>Lab Tools</name>
<location>right</location>
</webPart>
</preferredWebParts>
<permissions>
<permission>org.labkey.api.security.permissions.AdminPermission</permission>
</permissions>
</folderTab>
</folderTabs>
<modules>
<moduleName>Laboratory</moduleName>
</modules>
<menubarEnabled>true</menubarEnabled>
</folderType>
Tabbed Folders - The Active Tab
When creating a tabbed folder type, it is important to understand how the active tab is determined. The active tab is determined by the following checks, in order:
- If there is 'pageId' param on the URL that matches a tab's name, this tab is selected. This most commonly occurs after directly clicking a tab.
- If no URL param is present, the tabs are iterated from left to right, checking the selectors provided by each tab. If any one of the selectors from a tab matches, that tab is selected. The first tab with a matching selector is used, even if more than 1 tab would have a match.
- If none of the above are true, the left-most tab is selected
- View: This string will be matched against the viewName from the current URL (ie. 'page', from the current URL). If they are equal, the tab will be selected.
- Controller: This string will be matched against the controller from the current URL (ie. 'wiki', from the current URL). If they are equal, the tab will be selected.
- Regex: This is a regular expression that must match against the full URL. If it matches against the entire URL, the tab will be selected.
Modules: Query Metadata
If supplied, the metadata file should have the same name as the .sql file, but with a ".query.xml" extension (e.g., PeptideCounts.query.xml). For details on setting up the base query, see: Module SQL Queries.
For syntax details, see the following:
Examples
See the Examples section of the main query metadata topic.
The sample below adds table- and column-level metadata to a SQL query.
<query xmlns="http://labkey.org/data/xml/query">
<metadata>
<tables xmlns="http://labkey.org/data/xml">
<table tableName="ResultsSummary" tableDbType="NOT_IN_DB">
<columns>
<column columnName="Protocol">
<fk>
<fkColumnName>LSID</fkColumnName>
<fkTable>Protocols</fkTable>
<fkDbSchema>exp</fkDbSchema>
</fk>
</column>
<column columnName="Formulation">
<fk>
<fkColumnName>RowId</fkColumnName>
<fkTable>Materials</fkTable>
<fkDbSchema>exp</fkDbSchema>
</fk>
</column>
<column columnName="DM">
<formatString>####.#</formatString>
</column>
<column columnName="wk1">
<columnTitle>1 wk</columnTitle>
<formatString>####.#</formatString>
</column>
<column columnName="wk2">
<columnTitle>2 wk</columnTitle>
<formatString>####.###</formatString>
</column>
</columns>
</table>
</tables>
</metadata>
</query>
Metadata Overrides
Metadata is applied in the following order:
- JDBC driver-reported metadata.
- Module schemas/<schema>.xml metadata.
- Module Java code creates UserSchema and FilteredTableInfo.
- Module queries/<schema>/<query>.query.xml metadata.
- First .query.xml found in the active set of modules in the container.
- User-override query metadata within LabKey database, specified through the Query Schema Browser.
- First metadata override found by searching up container hierarchy and Shared container.
- For LABKEY.QueryWebPart, optional metadata config parameter.
Related Topics
Modules: Report Metadata
Example Report
Suppose you have a file-based R report on a dataset called "Physical Exam". The R report (MyRReport.r) is packaged as a module with the following directory structure.
externalModules
TestModule
queries
reports
schemas
study
Physical Exam
MyRReport.r
Report Metadata
To add metadata to the report, create a file named MyRReport.report.xml in the "Physical Exam" directory:
externalModules
TestModule
queries
reports
schemas
study
Physical Exam
MyRReport.r
MyRReport.report.xml
Using a metadata file, you can set the report as hidden, set the label and description, etc.
For details see the report metadata xml docs: ReportDescriptor.
A sample report metadata file. Note that label, description, and category are picked up by and displayed in the Data Views web part. Setting a report as hidden will hide it in Data Views web part and the Views menu on a data grid, but does not prevent the display of the report to users if the report's URL is called.
MyRReport.report.xml
<?xml version="1.0" encoding="UTF-8" ?>
<ReportDescriptor>
<label>My R Report</label>
<description>A file-based R report.</description>
<category>Reports</category>
<hidden>true</hidden>
</ReportDescriptor>
Modules: Custom Footer
To create a custom footer that appears on all pages throughout the site, place a file named _footer.html in your module, at the following location:
mymodule
resources
views
_footer.html
The footer can be written as an HTML fragment, without the <head> or <body> tags. The file can render any kind of HTML content, such as links, images, and scripts. It is also responsible for its own formatting, dependencies, and resources.
Images and CCS Files
Associated images and CSS files can be located in the same module, as follows:
mymodule
resources
web
mymodule
myimage.png
Example
The following _footer.html file references myimage.png.
<p align="center">
<img src="<%=contextPath%>/customfooter/myimage.png"/> This is the Footer Text!
</p>
Choosing Between Multiple Footers
If _footer.html files are contributed by multiple modules, you can select which footer to display from the Admin Console. Go to Admin > Site > Admin Console. Click Configure Footer.
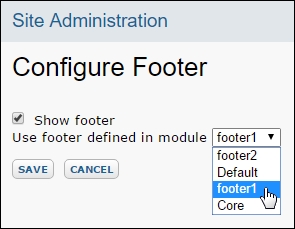
The dropdown list is populated by footers residing in modules deployed on the server (including both enabled and un-enabled modules).
- Core will show the standard LabKey footer "Powered by LabKey".
- Default will display the footer with the highest priority, where priority is determined by module dependency order. If module A depends on module B, then the footer in A has higher priority. Note that only modules that are enabled in at least one folder will provide a footer to the priority ranking process.
Modules: SQL Scripts
If your module is checked in to LabKey's subversion repository, or your module has the potential to be installed on additional servers (including by other developers), you should be especially conscious of updates to SQL scripts. Once a script has been checked in to LabKey's repository or run by another instance of LabKey, it is a good guideline to consider it immutable. If a table needs to be altered, no matter how trivial the change, a new upgrade script should normally be used. This is because if another server installs or upgrades using this script, it will not be re-run. If the script is then edited, this can cause the other machine to have an incomplete schema, which can easily result in errors downstream or on subsequent updates. For the case of scripts checked in to LabKey's subversion repository, be aware that other developers and LabKey's testing servers routinely run all checked-in scripts and it is very easy for problems to arise from inappropriately changed scripts. See the Hints and Advanced Topics section below for ways to make this process easier.
Note that module-based SQL scripts for assay types are not supported.
SQL Script Manager
You must name your SQL scripts correctly and update your module versions appropriately, otherwise your scripts might not run at all, scripts might get skipped, or scripts might run in the wrong order. The LabKey SQL Script Manager gets called when a new version of a module gets installed. Specifically, a module gets updated at startup time if (and only if) the version number listed for the module in the database is less than the current version in the code. The module version in the database is stored in core.Modules; the module version in code is returned by the getVersion() method in each Module class (Java module) or listed in version.properties (file-based module).
Rule #1: The module version must be bumped to get any scripts to run.
When a module is upgraded, the SQL Script Manager automatically runs the appropriate scripts to upgrade to the new schema version. It determines which scripts to run based on the version information encoded in the script name. The scripts are named using the following convention: <dBschemaName>-<fromVersion #.00>-<toVersion #.00>.sql
Rule #2: Use the correct format when naming your scripts; anything else will get ignored.
Use dashes, not underscores. Use two (or three, if required) decimal places for version numbers (0.61, 1.00, 12.10). We support three decimal places for very active modules, those that need more than 10 incremental scripts per point release. But most modules should use two decimal places.
Some examples:
- foo-0.00-1.00.sql: Upgrades foo schema from version 0.00 to 1.00
- foo-1.00-1.10.sql: Upgrades foo schema from version 1.00 to 1.10
- foo-1.10-1.20.sql: Upgrades foo schema from version 1.10 to 1.20
- foo-0.00-1.20.sql: Upgrades foo schema from version 0.00 to 1.20
The script directories can have many incremental & full scripts to address a variety of upgrade scenarios. The SQL Script Manager follows a specific algorithm when determining which script(s) to run for an upgrade. This is what it does:
- Determine installed module version number ("old") and new module version number ("new").
- Find all scripts in the directory that start at or above "old" and end at or below "new". Eliminate any scripts that have already been run on this database (see the core.SqlScripts table).
- Of these scripts, find the script(s) with the lowest "from" version. If there's just a single script with this "from" version, pick it. If there are more than one, pick the script with the highest "to" version.
- Run that script. Now the schema has been updated to the "to" version indicated in the script just run.
- Determine if more scripts need to be run. To do this, treat the "to" version of the script just run as the currently installed version.
- Repeat all the steps above (create list of scripts in the new range, eliminate previously run scripts, choose the script with the lowest starting point having the greatest range, and run it) until there are no more scripts left.
| Installed Module Version | New Module Version | Script(s) Run |
|---|---|---|
| 0.00 (not installed) | 1.10 | foo-0.00-1.00.sql, foo-1.00-1.10.sql |
| 0.00 (not installed) | 1.20 | foo-0.00-1.20.sql |
| 1.00 | 1.20 | foo-1.00-1.10.sql, foo-1.10-1.20.sql |
| 1.10 | 1.20 | foo-1.10-1.20.sql |
| 1.11 | 1.20 | None of these scripts |
Rule #3: Name your script as starting at the current module version in code.
This rule is important, but easily forgotten. If the most recent script in a directory is "foo-0.90-1.00.sql" and the new module version will be 2.00, it may be tempting to name the new script "foo-1.00-2.00.sql". This is almost certainly a mistake. What matters is the module version in code, not the ending version of the last script. The module number in code gets bumped for a variety of reasons (e.g., for a major release, for other schemas, or to force after-schema-update code to run), so a script that starts where the last script left off will probably never run. You must look at the current module version in code instead. There will be "gaps" in the progression; this is expected and normal.
If you're creating a new incremental script, here is a (nearly) foolproof set of steps that will produce a correct script name for module "Foo" that uses schema "foo":
- Finalize and test your script contents.
- Do an svn update to get current on all files. This ensures that no one else has bumped the version or checked in an incremental script with the same name.
- Find the current version number returned by the FooModule getVersion() method. Let's say it's 1.02.
- Name your script "foo-1.02-1.03.sql". (Incrementing by 0.01 gives you room to get multiple schema changes propagated and tested during the development period between major releases.)
- Bump the version number returned by FooModule.getVersion() to 1.03.
- Build, test, and commit your changes.
Rule #4: After a release, the next incremental script is still a point version of the release
Just before releasing a new version of LabKey Server, the LabKey team "rolls up" each module's incremental scripts into a single script for that release (e.g., foo-1.00-1.01.sql, foo-1.01-1.02.sql, and foo-1.02-1.03.sql get concatenated into foo-1.00-1.10.sql). This cleans things up a bit and reduces the number of script files, but it isn't required at all. The critical step is to get the incremental script right; you only get one chance for that.
The LabKey team will also bump all module versions to match the release. foo would now be version 1.10. The next script, intended for the 1.20 release, will be foo-1.10-foo-1.11.sql. Never bump the module version past the in-progress LabKey release. (e.g., if you get up to foo-1.18-1.19.sql before the 1.20 release, and still need another script, it would be foo-1.19-1.191.sql)
If you're testing an extensive schema change you may want to check in a script but not have it run on other developers' machines yet. This is simple; check in the script but don't bump the version number in code. When you're done testing, bump the version and everyone will upgrade.
The above guidelines eliminate most, but not all, problems with script naming. In particular, if multiple developers are working on the same module they must coordinate with each other to ensure scripts don't conflict with each other.
Remember that all scripts adhere to the module version number progression. If a single module manages multiple database schemas you must be extra careful about rule #3 and plan to see many gaps between each schema's script files.
Hints and Advanced Topics
- Modules are upgraded in dependency order, which allows schemas to safely depend on each other.
- Modules can (optionally) include two special scripts for each schema: <schema>-create.sql and <schema>-drop.sql. The drop script is run before all module upgrades and the create script is run after that schema's scripts are run. The primary purpose is to create and drop SQL views in the schema. The special scripts are needed because some databases don't allow modifying tables that are used in views. So LabKey drops all views, modifies the schema, and re-creates all views on every upgrade.
- Java upgrade code. Some schema upgrades require code. One option is to implement and register a class in your module that implements UpgradeCode and invoke its methods from inside a script via the core.executeJavaUpgradeCode stored procedure. This works well for self-contained code that assumes a particular schema structure; the code is run once at exactly the right point in your upgrade sequence.
- After schema update. Another option for running Java code is to call it from the Module afterUpdate() method. This can be useful if the upgrade code needs to call library methods that change based on the current schema. Be very careful here; the schema could be in a completely unknown state (if the server hasn't upgraded in a while then your code could execute after two years of future upgrade scripts have run).
- ant bootstrap. On a developer machine: shut down your server, run "ant bootstrap", and restart your server to initiate a full bootstrap on your currently selected database server. This is a great way to test SQL scripts on a clean install. Use "ant pick_pg" and "ant pick_mssql" to test against the other database server.
- The Admin Console provides other helpful tools. The "Sql Scripts" link shows all scripts that have run and those that have not run on the current server. From there, you can choose to "Consolidate Scripts" (e.g., rolling up incremental scripts into version upgrade scripts or creating bootstrap scripts, <schema>-0.00-#.00.sql). While viewing a script you have the option to "Reorder" the script, which attempts to parse and reorder all the statements to group all modifications to each table together. This can help streamline a script (making redundant or unnecessary statements more obvious), but is recommended only for advanced users.
- In addition to these scripts, you will need to create a schema XML file. This file is located in the /scripts folder of your module. There is one XML file per schema. This file can be auto-generated for an existing schema. To get an updated XML file for an existing schema, go to the Admin Console then pick 'Check Database'. There will be a menu to choose the schema and download the XML. If you would like to download an XML file for a schema not yet visible to labkey, you can use a URL along these lines directly: http://localhost:8080/labkey/admin/getSchemaXmlDoc.view?dbSchema=<yourSchemaName>. Simply replace the domain name & port with the correct values for your server. Also put the name of your schema after 'dbSchema='. Note: Both the schema XML file name and 'dbSchema=' value are case-sensitive. They must match the database schema name explicitly.
- LabKey offers automated tests that will compare the contents of your schema XML file with the actual tables present in the DB. To run this test, visit a URL similar to: http://localhost:8080/labkey/junit/begin.view?, but substitute the correct domain name and port. Depending on your server configuration, you may also need to omit "/labkey" if labkey is run as the root webapp. This page should give a list of all Junit test. Run the test called "org.labkey.core.admin.test.SchemaXMLTestCase".
- Schema delete. When developing a new module, schemas can change rapidly. During initial development, it may be useful to completely uninstall / reinstall a module in order to rebuild the schema from scratch, rather than make changes via a large number of incremental scripts. Uninstalling a module requires several steps: drop the schema, delete the entry in the core.Modules table, delete all the associated rows in the core.SqlScripts table. The "Module Details" page (from the Admin Console) provides a quick way to uninstall a module; when your server is restarted, the module will be reinstalled and the latest scripts run. Use extreme caution… deleting a schema or module should only be done on development machines. Also note that while this is useful for development, see warnings above about editing scripts once checked into subversion and/or otherwise made available to other instances of LabKey.
Script Conventions
The conventions below are designed to help everyone write better scripts. They 1) allow developers to review & test each other's scripts and 2) produce schema that can be changed easily in the future. The conventions have been developed while building, deploying, and changing production LabKey installations over the last eight years; we've learned some lessons along the way.
Databases & Schemas
Most modules support both PostgreSQL and Microsoft SQL Server. LabKey Server uses a single primary database (typically named "labkey") divided into 20 - 30 "schemas" that provide separate namespaces, usually one per module. Note that, in the past, SQL Server used the term "owner" instead of "schema," but that term is being retired.
Capitalization
SQL keywords should be in all caps. This includes SQL commands (SELECT, CREATE TABLE, INSERT), type names (INT, VARCHAR), and modifiers (DEFAULT, NOT NULL).
Identifiers such as table, view, and column names are always initial cap camel case. For example, ProtInfoSources, IonPercent, ZScore, and RunId. Note that we append 'Id' (not 'ID') to identity column names.
We use a single underscore to separate individual identifiers in compound names. For example, a foreign key constraint might be named 'FK_BioSource_Material'. More on this below.
Constraints & Indexes
Do not use the PRIMARY KEY modifier on a column definition to define a primary key. Do not use the FOREIGN KEY modifier on a column definition to define a foreign key. Doing either will cause the database to create a random name that will make it very difficult to drop or change the index in the future. Instead, explicitly declare all primary and foreign keys as table constraints after defining all the columns. The SQL Script Manager will enforce this convention.
- Primary Keys should be named 'PK_<TableName>'
- Foreign Keys should be named 'FK_<TableName>_<RefTableName>'. If this is ambiguous (multiple foreign keys between the same two tables), append the column name as well
- Unique Constraints should be named 'UQ_<TableName>_<ColumnName>'
- Normal Indexes should be named 'IX_<TableName>_<ColumnName>'
- Defaults are also implemented as constraints in some databases, and should be named 'DF_<TableName>_<ColumnName>'
Keep Your SQL as Database-Independent as Possible
You may prefer using PostgreSQL over SQL Server (or vice versa), but don't forget about the other database… write your scripts to work with both databases and you'll save yourself many headaches. Test your scripts on both databases.
Statement Endings
Every statement should end with a semicolon, on both PostgreSQL and SQL Server. In older versions of SQL Server, "GO" statements needed to be interjected frequently within SQL scripts. They are rarely needed now, except in a few isolated cases:
- After creating a new user-defined type (sp_addtype), which is rare
- Before and after a stored procedure definition; SQL Server requires each stored procedure definition to be executed in its own block
- After a DROP and re-CREATE
- After an ALTER statement, if the altered object is referenced later in the scripts
Scripting from SQL Server
It is often convenient to create SQL Server objects or data via visual tools first, and then have SQL Server generate the correct CREATE, INSERT, etc scripts. This is fine; however be aware that the script will have a "USE database name" statement at the top. Be sure to remove this before committing your upgrade script, as the database name in other environments is entirely arbitrary.
Related Topics
Modules: Database Transition Scripts
Modules that need to store their own data may find it useful to create a new schema and set of related tables in the relational database used by LabKey Server. Modules can transition schemas between versions by including database transition scripts.
Generate a schema
You can generate a basic version of the schema file for an existing schema by navigating to a magic URL:
http://<server>/labkey/admin/getSchemaXmlDoc.view?dbSchema=<schema-name>
Save the result to the /schema/<schema-name>.xml file in your module.
Store schema transition scripts
Schema transition scripts should live in the schemas/dbscripts/<db-type>/ directory of your module. Currently, the following database types are supported:
| Database Type | Directory |
|---|---|
| PostgreSQL | schemas/dbscripts/postgresql/ |
| Microsoft SQL Server | schemas/dbscripts/sqlserver/ |
The name of the script is also quite important. Each script in this directory moves the database schema from one version of your module to another. The name of the file indicates which versions the script will transition from and to. The general format is <schemaname>-<oldversion>-<newversion>.sql. For more details about how these scripts work, see Modules: SQL Scripts.
For example, to create a new schema with some tables for your module (which we have assigned a version number of 1.0) on a PostgreSQL database, you would create a new SQL script file in the following location:
externalModules/resources/schemas/dbscripts/postgresql/ReportDemo-0.0-1.0.sql
Your schema name can be anything that does not conflict with any existing schema name, so it's generally best for your schema to be named the same as your module.
When a new version of a module appears, the server will restart and, during its initialization, it will execute any relevant database scripts. Once the scripts to bring the module to version 1.0 have been executed, the module will report its version as 1.0, and those scripts will not be run again. If you need to make changes to your database schema, adjust your module version to 1.1, and create a new SQL script to transition the database schema from version 1.0 to 1.1. The file name for that would be:
externalModules/resources/schemas/dbscripts/postgresql/ReportDemo-1.0-1.1.sql
Related Topics
See Modules: SQL Scripts, which describes these files in detail.
Modules: Domain Templates
Domain templates are currently "experimental" - the file formats or behavior may change in upcoming releases.
A domain template is an xml file that can be included in a module that specifies the shape of a Domain, for example, a List, SampleSet, or DataClass. An example template xml file can be found in our test module:
test/modules/simpletest/resources/domain-templates/todolist.template.xml - link to source
A domain template includes:
- a name
- a set of columns
- an optional set of indices (to add a uniqueness constraint)
- an optional initial data file to import upon creation
- domain specific options (e.g, for SampleSet, the list of columns that make the Name column unique.)
While not present in the domainTemplate.xsd, a column in a domain template can be marked as "mandatory". The domain editor will not allow removing or changing the name of mandatory columns. For example,
<templates
xmlns="http://labkey.org/data/xml/domainTemplate"
xmlns:dat="http://labkey.org/data/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance">
<template xsi:type="ListTemplateType">
<table tableName="Category" tableDbType="NOT_IN_DB" hidden="true">
<dat:columns>
<dat:column columnName="category" mandatory="true">
<dat:datatype>varchar</dat:datatype>
<dat:scale>50</dat:scale>
</dat:column>
</dat:columns>
</table>
<initialData>
<file>/data/category.tsv</file>
</initialData>
<options>
<keyCol>category</keyCol>
</options>
</template>
</templates>
All domains within in a template group can be created from the template via JavaScript API:
LABKEY.Domain.create({
domainGroup: "todolist",
importData: false
});Or a specific domain:
LABKEY.Domain.create({
domainGroup: "todolist",
domainTemplate: "Category",
importData: false
});When "importData" is false, the domain will be created but the initial data won't be imported. The importData flag is true by default.
When "createDomain" is false, the domain will not be created, however any initial data will be imported.
A domain template typically has templates with unique names, but it is possible to have a template with the same name of different domain kinds -- for example, a DataClass template and a SampleSet template named "CellLine". In this situation, you will need to disambiguate which template with a "domainKind" parameter. For example,
LABKEY.Domain.create({
domainGroup: "biologics",
domainTemplate: "CellLine",
domainKind: "SampleSet",
createDomain: false,
importData: true
});Deploy Modules to a Production Server
Typically you will develop a module on a test server and then move it to the production server once the development is complete. Moving the module can be done either by copying the uncompressed module directory and its subdirectories and files from the test server to the production server, or by compressing the module directory into a .module file and copying that to the production server. Which technique you choose will probably depend on what kind of file system access you have between the servers. If the production server's drive is mounted on the test server, a simple directory copy would be sufficient. If FTP is the only access between the test and production servers, sending a compressed file would be easier.
An easy way to compress the module directory is to use the JAR utility, which can also be automated via an ANT build script. Use the standard JAR options and name the target file "<module-name>.module".
Deploy the .module file to the <labkey-home>/externalModules/ directory on your production server. A server running in production mode will not recognize a new module -- a manual server restart is required in this case. But a production server will monitor existing modules for changes. When it loads the module, it will automatically expand the .module into a directory with the same base name (overwriting the existing directory and files), and load the newly-updated module's resources. Also a module must be enabled in the folder you are working/testing in.
Most files in a module can be updated while the production server is running (sql queries, html views, trigger scripts, assay domains and views) without restarting. Some files cannot be updated while the server is running (SQL scripts, assay provider definitions, compiled Java code, etc) and require a manual restart of the server.
Related Topics
Upgrade Modules
When launching Tomcat on the command line, you can point the server at a different externalModules directory using the externalModulesDir parameter:
-Dlabkey.externalModulesDir=/MY/OTHER/DIRECTORY
It is important to note that LabKey Server does not provide binary compatibility between releases. Therefore, before upgrading a production installation with custom modules, you must first ensure that your custom modules build and operate correctly with the new version of the server. Deploying a module written for a different version of the server will have unpredictable and likely undesirable results.
Main Credits Page
Modules can contribute content to the main credits page on your LabKey Server.
To add a credits page to your module, create a jars.txt file documenting all jars and drop it in the following directory: <YOUR MODULE DIRECTORY>\src\META-INF\<YOUR MODULE NAME>.
The jars.txt file must be written in wiki language and contain a table with appropriate columns. See the following example:
{table}
Filename|Component|Version|Source|License|LabKey Dev|Purpose
annotations.jar|Compiler
annotations|1.0|{link:JetBrains|http://www.jetbrains.com/}|{link:Apache
2.0|http://www.apache.org/licenses/LICENSE-2.0}|adam|Annotations to enable compile-time checking for null antlr-3.1.1.jar|ANTLR|3.1.1|{link:ANTLR|http://www.antlr.org/}|{link:BSD|http://www.antlr.org/license.html}|mbellew|Query
language parsing
axis.jar|Apache
Axis|1.2RC2|{link:Apache|http://ws.apache.org/axis/}|{link:Apache
2.0|http://www.apache.org/licenses/LICENSE-2.0}|jeckels|Web service implementation
{table}
Module Properties Reference
myModule
│ module.properties
└───resources
├───...
├───...
└───...
The following module.properties file is for a simple file-based module which contains no Java classes to compile:
Name: HelloWorld
ModuleClass: org.labkey.api.module.SimpleModule
Version: 1.0
Modules that contain Java classes should reference their main Java class. For example, the Issues module references the main controller class org.labkey.issue.IssuesModule:
ModuleClass: org.labkey.issue.IssuesModule
ModuleDependencies: Wiki, Experiment
Label: Issue Tracking Service
Description: The LabKey Issues module provides an issue tracker, a centralized workflow system for tracking issues or tasks across the lifespan of a project. Users can use the issue tracker to assign tasks to themselves or others, and follow the task through the work process from start to completion.
URL: https://www.labkey.org/wiki/home/Documentation/page.view?name=issues
Organization: LabKey
OrganizationURL: https://www.labkey.com/
License: Apache 2.0
LicenseURL: http://www.apache.org/licenses/LICENSE-2.0
Properties Reference
Available properties for modules. Note that property names vary slightly between module.property and module.xml files.
| Property Name (in module.xml) | Property Name (in module.properties) | Description |
|---|---|---|
| class | ModuleClass | Main class for the module. For modules without Java code, use org.labkey.api.module.SimpleModule |
| name | Name | The display name for the module. |
| version | Version | The module version. |
| requiredServerVersion | RequiredServerVersionooo | The minimum required version for LabKey Server. |
| moduleDependencies | ModuleDependencies | A comma-delimited list of other module names this module depends upon. This determines module initialization order and controls the order in which SQL scripts run. For example, suppose your module includes a foreign key to a table in the Experiment module. In this case you could declare a dependency on the Experiment module, so that you can be sure that the target table exists before you try to create your foreign key. LabKey Server will give an error if you reference a module that doesn't exist, or if there's a circular dependency, for example, if ModuleA depends on ModuleB, which itself depends on ModuleA. |
| supportedDatabases | SupportedDatabases | Add this property to indicate that your module runs only on a particular database. Possible values: "pgsql" or "mssql". |
| label | Label | One line description of module's purpose (display capitalized and without a period at the end). |
| description | Description | Multi-line description of module. |
| url | URL | The homepage URL for additional information on the module. |
| author | Author | Comma separated list of names and, optionally, email addresses: e.g. "Adam Rauch <adamr@labkey.com>, Kevin Krouse" |
| maintainer | Maintainer | Comma separated list of names and, optionally, email addresses: e.g. "Adam Rauch <adamr@labkey.com>, Kevin Krouse" |
| organization | Organization | The organization responsible for the module. |
| organizationURL | OrganizationURL | The organization's URL/homepage. |
| license | License | License name: e.g. "Apache 2.0", "GPL-2.0", "MIT" |
| licenseURL | LicenseURL | License URL: e.g. "http://www.apache.org/licenses/LICENSE-2.0" |
| vcsRevision | VcsRevision | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The SVN revision number of the module. This will be displayed next to the module in the site admin console. |
| vcsUrl | VcsURL | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The URL to the SVN server that manages the source code for this module. This will be displayed next to the module in the site admin console. |
| buildOS | BuildOS | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The operating system upon which the module was built. This will be displayed next to the module in the site admin console. |
| buildPath | BuildPath | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The file path in which the module was built. This will be displayed next to the module in the site admin console. |
| buildTime | BuildTime | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The date and time the module was built. This will be displayed next to the module in the site admin console. |
| buildType | BuildType | Possible values are "Development" or "Production". "Development" modules will not deploy on a production machine. To build modules destined for a production server, run 'ant production', or add the following to your module.properties file: 'BuildType=Production'. |
| buildUser | BuildUser | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The name of the user that built the module. This will be displayed next to the module in the site admin console. |
| sourcePath | SourcePath | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The location of the module source code. |
| resourcePath | ResourcePath | This value is set internally by the build, and does not need to be provided by the developer in module.properties. |
| buildNumber | BuildNumber | This value is set internally by the build, and does not need to be provided by the developer in module.properties. The build number. |
| enlistmentId | EnlistmentId | This value is set internally by the build, and does not need to be provided by the developer in module.properties. Used to determine whether the module was built on the current server. |
Properties Surfaced in the Admin Console
Module properties are surfaced in the user interface at Admin > Site > Admin Console, under the heading Module Information. Click an individual module name to see its properties. If you having problems loading/reloading a module, check the properties Enlistment ID and Source Path. When the server is running in devMode, these properties are displayed in green text if the values in module.xml match the values found the on the server; they are displayed in the red text if there is a mismatch.
The properties for deployed modules are available in the table core.Modules, where they can be accessed by the client API.
Generation of module.xml
When you run the standard Ant build targets in the open source project, the property/value pairs in module.properties are extracted and used to populate a module.xml file (via string substitution into module.template.xml). The resulting module.xml file is copied to the module's config subdirectory (MODULE_NAME/config/module.xml) and finally packaged into the built .module file. At deployment time, the server loads properties from config/module.xml, not module.properties (which the server ignores). Note that modules that contain Java code must be built using the standard build targets in the open source project.
Note that there are two different files named "module.xml" inside a module's source code: the one located at MODULE_NAME/module.xml is for declaring dependencies on libraries and other resources; the one located at MODULE_NAME/config/module.xml is generated based on module.properties and is for setting module properties.
An example module.xml file generated from module.properties:
<?xml version="1.0" encoding="UTF-8"?>
<beans xmlns="http://www.springframework.org/schema/beans"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xmlns:util="http://www.springframework.org/schema/util"
xsi:schemaLocation="http://www.springframework.org/schema/beans http://www.springframework.org/schema/beans/spring-beans-2.0.xsd">
<bean id="moduleBean" class="org.labkey.api.module.SimpleModule">
<property name="name" value="mymodule"/>
<property name="version" value="0.0"/>
<property name="requiredServerVersion" value="0.0"/>
<property name="moduleDependencies" value="Wiki"/>
<property name="label" value="My Module"/>
<property name="description">
<value><![CDATA[My module helps users solves problems related to...]]></value>
</property>
<property name="url" value="https://www.mywebsite.com"/>
<property name="author">
<value><![CDATA[Jane Doe <janedoe@email.com>]]></value>
</property>
<property name="maintainer">
<value><![CDATA[John Doe <johndoe@email.com>]]></value>
</property>
<property name="organization" value="My Software Inc."/>
<property name="organizationUrl" value="https://www.my-software-inc.com/"/>
<property name="license" value="Apache 2.0"/>
<property name="licenseUrl" value="http://www.apache.org/licenses/LICENSE-2.0"/>
</bean>
</beans>