Table of Contents |
guest 2025-05-30 |
Register Molecules
Molecular Physical Property Calculator
Molecules, Sets, and Molecular Species
Definitions
Molecules are composed of various components and have a set of physical properties that describe them. When a molecule is added to the registry, the following additional entities are calculated and added, depending on the nature of the molecule. These additional entities can include:
- Molecular Species
- Molecule Sets
- Other Protein Sequences
Molecular Species serve as alternate forms for a given molecule. For example, a given antibody may give rise to multiple Molecular Species: one Species corresponding to the leaderless, or "mature", portion of the original antibody and another Molecular Species corresponding to its "mature, desK" (cleaved of signal peptides and heavy chain terminal lysine) form.
Both Molecular Species and Sets are calculated and created by the registry itself. Their creation is triggered when the user adds a Molecule to the registry. Users can also manually register Molecular Species, but generally do NOT register their own Molecule Sets. Detailed triggering and creation rules are described below.
Molecule Components
A molecule can be created (= registered) based on one or more of the following:
- a protein sequence
- a nucleotide sequence
- other molecules
Rules for Entity Calculation/Creation
When the molecule contains only protein sequences, then:
- A mature molecular species is created, consisting of the leaderless segments of the protein sequences, provided a leader portion is identifiable. The leader segment has to be:
- Have annotation start with residue #1
- The annotation Type is "Leader"
- The annotation Category is "Region"
- (If no leader portion is identifiable, then the species will be identical to the Molecule which has just been created.)
- Additionally, a mature desK molecular species is created (provided that there are terminal lysines on heavy chains).
- New protein sequences are created corresponding to any species created, either mature, mature des-K, or both. The uniqueness constraints imposed by the registry are in effect, so already registered proteins will be re-used, not duplicated.
- A molecule set is created, provided that the mature molecular species is new, i.e., is not the same (components and sequences) as any other mature molecular species of another molecule. If there is an already existing mature species in the registry, then the new molecule is associated with that set.
- Physical properties are not calculated.
- Molecular species are not created.
- A molecule set is created, which has only this molecule within it.

Aliases and Descriptions
When creating a Molecule, the auto-generated Molecule Set (if there is a new one) will have the same alias as the Molecule. If the new Molecule is tied to an already existing Molecule Set, the alias is appended with alias information from the new Molecule.
When creating a molecule, the auto-generated molecular species (both mature and mature desK) should have the same alias as the molecule. Similarly, when creating a molecular species from a molecule (manually), the alias field will pre-populate with the alias from the molecule.
For molecular species that are auto-generated, if it is creating new protein sequences (one or more) as the components of that molecular species, they have a Description:
- Mature of “PS-15”
- Mature, desK of “PS-16”
- Mature of “PS-17”
- Mature, desK of “PS-18”
Related Topics
Register Molecules
This topic shows how to register a new molecule using the graphical user interface. To register molecules in bulk via file import, see Create Registry Sources.
Other ways to register molecules are to:Create a Molecule
To add a new molecule to the registry:
- Open the Molecules creation wizard.
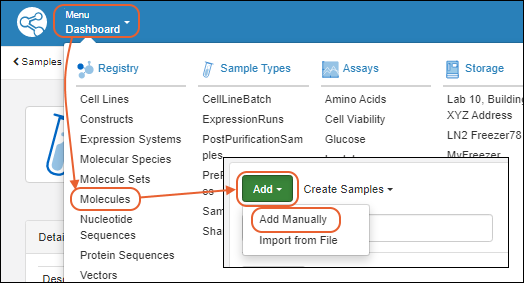
Add Details
On the first tab of the wizard, enter the following:
- Name: Provide a name, or one will be generated for you. Hover to see the naming pattern
- Description: (Optional) A text description of the molecule.
- Alias: (Optional) Alternative names for the molecule.
- Common Name: (Optional) The common name of the molecule, if any.
- Molecule Parents: (Optional) Parent molecules for the new molecule.
Click Next to continue.
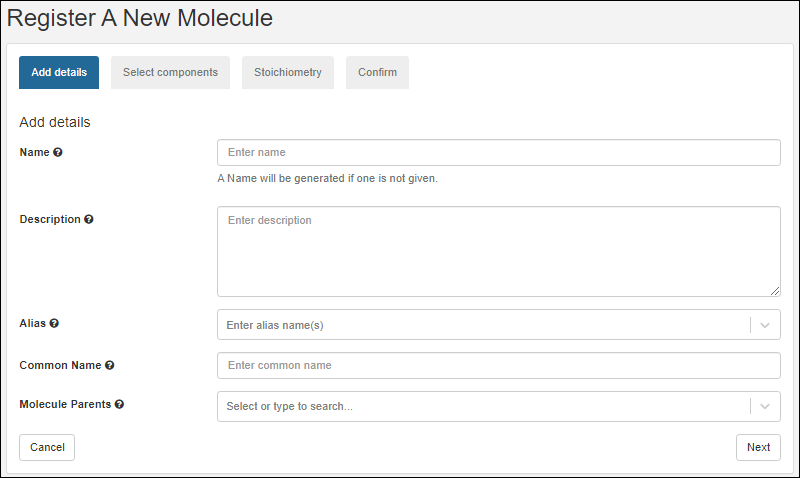
Select Components
- On the Select components tab, search and select existing components of the new molecule.
- After selecting the appropriate radio button, search for the component of interest.
- Type ahead to narrow the list.
- You will see a details preview panel to assist you.
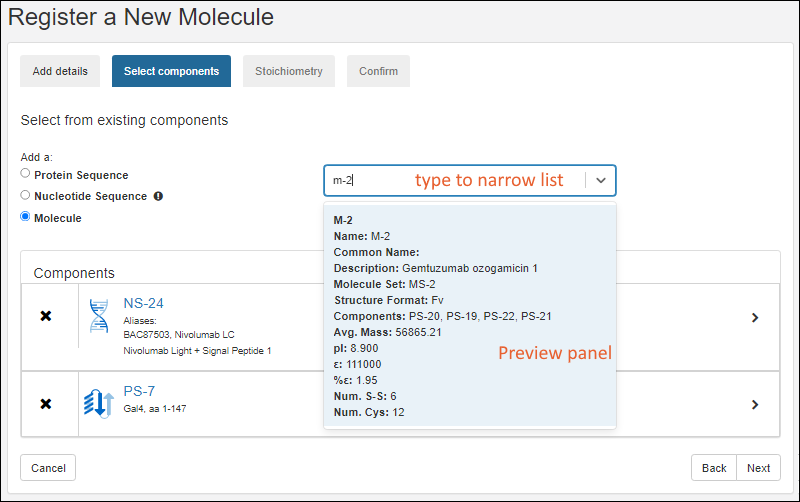
- Once you have added a component, it will be shown as a panel with entity icon. Click the to expand details.
Click Next to continue.
Stoichiometry
LabKey Biologics will attempt to classify the structure format of the molecule's protein components, if possible. The structure format is based on the component protein chain formats.
On the Stoichiometry pane enter:
- Stoichiometry from each component
- Structure Format: Select a format from the pulldown list. The list is populated from the StructureFormat table.
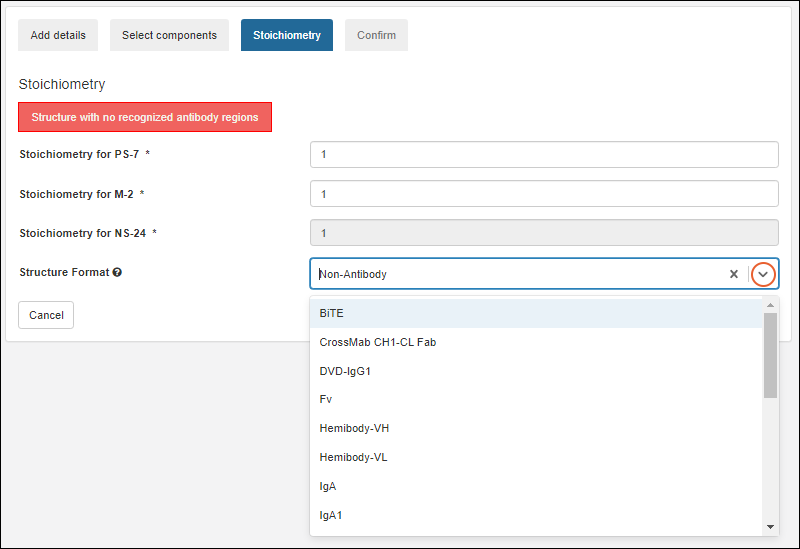
Click Next to continue.
Confirm
On the final tab, confirm the selections and click Finish to add the molecule to the registry.
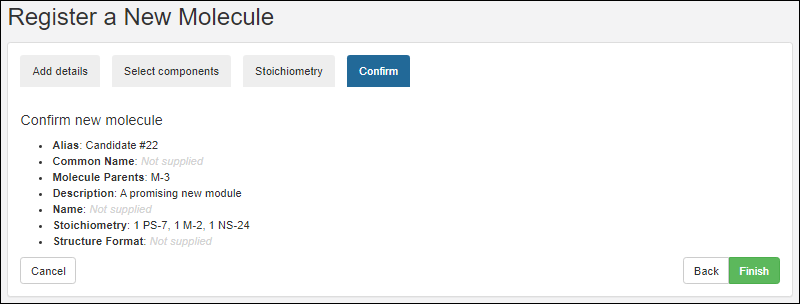
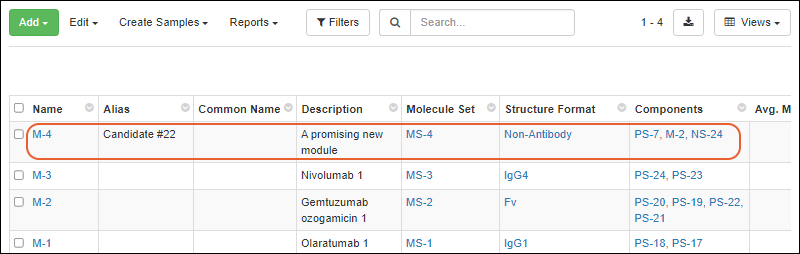
The new molecule will be added to the grid.
Related Topics
Molecular Physical Property Calculator
Antibody discovery, engineering, and characterization work involves a great deal of uncertainty about the materials at hand. There are important theoretical calculations necessary for analysis as well as variations of molecules that need to be explored. Scientists want to consider and run calculations several different ways based on variations/modifications they are working with for analysis and inclusion in a notebook.
In the case of a structure format not being recognized, e.g. some scFv diabody, it will not be properly classified and the calculations will be wrong. Providing the ability to reclassify and recalculate molecular physical properties is key for assisting with scenarios such as "What if this S-S bond formed or didn’t?"
Using the built-in Molecular Physical Property Calculator, you can view the persisted calculations to make your input conditions and calculation type clearer and select alternative conditions and calculations for your entity.
We currently calculate average mass, pI (isoelectric point), and 𝜺 (extinction coefficient) from the sequence, molecular stoichiometry, number of free Cysteine and disulfide bonds. While including stoichiometry, free Cys vs S-S, these inputs include sequence scope/type.
View Physical Property Calculator
From the grid of Molecules, select the Molecule of interest. Click the Physical Property Calculator tab.
Calculation Inputs
In the Calculation Inputs panel, you'll enter the values to use in the calculation:
- Num. S-S: Dynamically adjusted to be half of the "Num. Cys" value.
- Num Cys: A display only value derived from the sequence chosen.
- Sequence Scope: Select the desired scope. Sequence ranges will adjust based on your selection of any of the first three options. Use "Custom Range" for finer control of ranges.
- Full Protein Sequence: complete amino acid sequence of a protein, including all the amino acids that are synthesized based on the genetic information.
- Mature Protein Sequence: final, active form of the protein, after post-translational modifications and cleavages, or other changes necessary for the protein to perform its intended biological function.
- Mature Des-K Protein Sequence: mature protein sequence with the C-Terminal Lysine removed.
- Custom Range
- Sequence Ranges for each component sequence. These will adjust based on the range selected using radio buttons, or can be manually set using the "Custom Range" option.
- Use Range: Enter the start and end positions.
- Click View this sequence to open the entire annotated sequence in a new tab for reference.
- Stoichiometry
- Analysis Mode. Select from:
- Native
- Reduced
- Alkylated Cysteine (only available in "Reduced" mode). If available, select the desired value.
- Modifiers
- Pyro-glu: Check the box for "Cyclize Gln (Q) if at the N-terminus"
- PNGase: Check the box for "Asn (N) -> Asp (D) at N-link sites"
Properties
When you click Calculate, the right hand panel of Properties will be populated. You'll see both what the Classifier Generated value is (if any) and the Simulated value using the inputs you entered.
Calculations are provided for:
- Mass
- Average Mass
- Monoisotopic Mass
- Organic Average Mass
- pI - Isoelectric Point calculated by different methods:
- Bjellqvist
- EMBOSS
- Grimsley
- Patrickios (simple)
- Sillero
- Sillero (abridged)
- Other
- Chemical Formula
- Extinction Coefficient - ε
- Percent Extinction Coefficient
- Sequence Length
- Amino Acid (AA) composition
Export Calculations
To export the resulting calculated properties, click Export Data.
The exported Excel file includes both the calculated properties and the inputs you used to determine them. For example, the export of the above-pictured M-17 calculation would look like this: