File Watchers let administrators
set up the monitoring of directories on the file system and perform specific actions when desired files appear. This topic describes the various pipeline tasks available. The configuration options for file watcher triggers and specifics about type of files eligible vary based on the type of file watcher, and the tasks available vary by folder type.
File Watcher Tasks
Reload Folder Archive (Any folder)
This option is available in any folder type and reloads an unzipped folder archive. It will not accept compressed (.zip) folder archives.
The reloader expects a folder.xml in the base directory of the archive. To create an unzipped folder archive,
export the folder to your browser as a .zip file and then unzip it.
Reload Lists Using Data File (Any folder)
This option is available in any folder type and imports data to existing lists from source files. It can also infer non-key column changes. Note that this task cannot create a new list definition: the list definition must already exist on the server. The list module must also be enabled in the folder.
Move Files Across the Server (Any folder)
This option is available in any folder type. It moves and/or copies files around the server without analyzing the contents of those files.
Import/Reload Study Datasets Using Data File (Study folders only)
This option is available in a study folder. It loads data into existing study datasets and it infers/creates datasets if they don't already exist. Source data can be in TSV, Excel, or text files.
Reload Study (Study folders only)
This option reloads or populates a study from an unzipped study archive. This task is triggered by a .txt file (i.e. studyload.txt) and reloads a study (both its datasets and study configurations such as cohort assignments). To create a study archive, see
Export a Study.
Imports Specimen Data Using Data File (Study folders only)
This file watcher type accepts specimen data in both .zip and .tsv file formats:
- .zip: The specimen archive zip file has a .specimens file extension.
- .tsv: An individual specimens.tsv file which will typically be the simple specimen format and contain only vial information. This file will have a # specimens comment at the top.
By default, specimen data imported using the a file watcher will be set to
replace existing data. To
merge instead, set the property "mergeSpecimen" to true.
Import a Directory of FCS Files (Flow folders only)
Import flow files to the flow module. This type of file watcher is only available in Flow folders. It supports a process where FCS flow data is deposited in a common location by a number of users, with each data export placed into a subdirectory of the watched folder, perhaps in a separate subdirectory per user.
When the File Watcher finds these files, they are placed into a new location under the folder pipeline root based on the current user and date. Example: @pipeline/${username}/${date('YYYY-MM')}
LabKey then imports the FCS data to that container. All FCS files within a single directory are imported as a single experiment run in the flow module.
Custom Parameters
Custom parameters are available as follows:
| Custom Parameter | Available for 'Reload lists'? | Available for 'Import/reload study datasets'? | Available for 'Imports specimen data...'? | Default Value |
|---|
| mergeData | Yes | No | No | False (replace) |
| mergeSpecimen | No | No | Yes | False (replace) |
| default.action | No | Yes | No | Replace |
| allowDomainUpdates | Yes | Yes | No | True |
mergeData
This parameter is available for the task 'Reload Lists Using Data File'.
By default, the 'Reload Lists' task replaces list data using the contents of the source files. You may merge data by including the custom parameter: "mergeData": "true".

mergeSpecimen
This parameter is available for the task 'Imports specimen data using data file'.
By default, the specimen import task replaces specimen data using the contents of the import. You may merge data by including the custom parameter: "mergeSpecimen": "true".

default.action
This parameter is available for the task 'Import/Reload Study Datasets Using Data File'.
For study datasets, the parameter 'default.action' controls whether incoming data is appended to existing datasets or replaces the datasets entirely. If you do not include this parameter, or set it to 'replace', the incoming data will replace all existing data in the dataset. To keep the current dataset and append incoming data, set the parameter to 'append'.

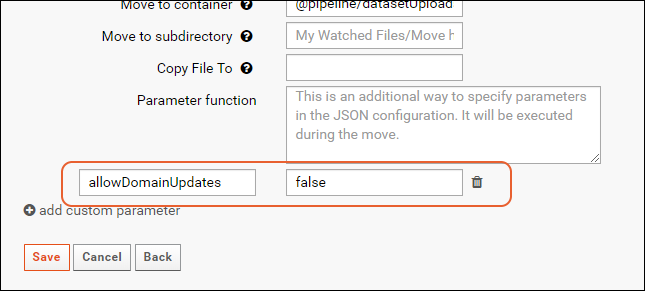
allowDomainUpdates
This parameter is available for the tasks 'Reload Lists Using Data File' and 'Import/Reload Study Datasets Using Data File'.
When updating lists and datasets, by default, the columns in the incoming data will overwrite the columns in the existing list or dataset. This means that any new columns in the incoming data will be added to the list and any columns missing from the incoming data will be dropped (and their data deleted).
To override this behavior, set the parameter "allowDomainUpdates" to false to retain the column set of the existing list.
Related Topics
