Search
LabKey v10.1 introduces a new search engine that provides full-text search across your server using
Lucene search syntax. Search is secure, so you only see results that you have sufficient permissions to view. Results are ordered by relevance. For details, please see LabKey's search documentation.
Participant Searches. Study managers are often particularly interested in seeing all of the data available for a particular individual across all studies. It is now easy to search for a particular participant ID. The appropriate participant page will be the top hit, followed by attachments and other documents that mention this participant.
Example. Searching labkey.org for participant 249318596 shows
these results. The
participant page for this individual tops the list, followed by files that include this participant and several pages of documentation. Security rules ensure that only public studies are included in results.
Data types. Most data types on your server are searched:
- Metadata for datasets and assays.
- Includes column names for datasets
- Includes descriptions, types, etc for assays.
- Participant IDs
- Wikis, including attachments
- Messages, including attachments
- Issues
- Files
- Automatically includes the contents of all file directories.
- Does not include the contents of a pipeline override unless this location allows search.
- Does not include the contents of .zip archives.
File Upload
File management has become more powerful, centralized and streamlined in LabKey v10.1. Please see the
file documentation for full coverage of 10.1 improvements, which include:
- Easy upload of groups of files. You can now upload entire directories of files or multiple individual files without installing a separate WebDav client for multi-file management.
- No pipeline management required. Default settings for file storage now allow most admins to skip setting up a pipeline (now called a pipeline override).
- Assay file upload can be centralized. The "Files" web part now provides a central location for file processing for all different types of assays.
- Support for a two-step, upload-then-import assay workflow. It is now easy to separate the task of uploading many assay files to your server from the process of importing these files into assay data structures. See details on your options in the picture below.
Illustration. LabKey v10.1 introduces a a new, two-step alternative for bringing data into the LabKey database:
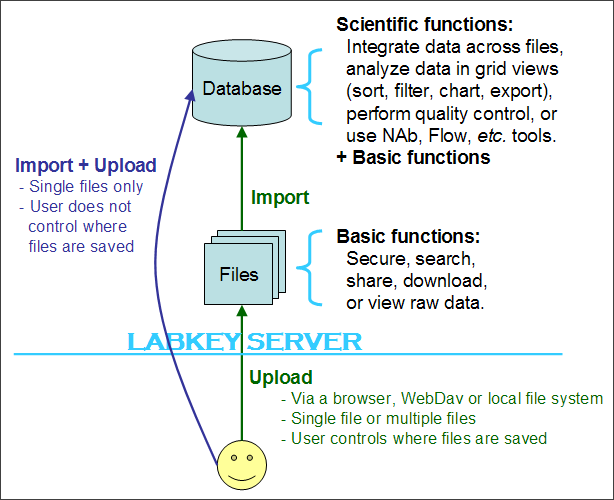 Upgrade Tips
Upgrade Tips. The enhancements mentioned above required major changes to the locations where files are stored on the file system. Tips:
- Selection of a site-level file root occurs during upgrade. During upgrade, if you have not previously specified a site-level file root, you will be prompted to enter one. You can either accept the default or override with a new one.
- Files are migrated during upgrade.
- During upgrade, the system looks in all existing file roots and moves the files it finds into new @files directories in a new directory structure that mirrors the project/folder structure of your LabKey Server. The location of the @files directories (and thus the destination of each set of files) is determined by the site-level file root you set at upgrade time. Note that file roots were called web roots in 9.3.
- Of the three different types of directory roots that LabKey supports (file sets, pipeline, and named file sets), this change only affects file sets. Pipeline and named file sets are considered 'external files' and are not managed by the server. If you don't know what a named file set is, you most likely do not have any, so you can ignore this tip.
- You can check to see which files may be affected by looking at the files summary view (linked on the admin console). This page shows all three types of roots. Only those under the @files node are affected. Logging (to the console) is in place to display movement of files and any errors.
- If you have a pipeline root and a web root set to the same location and you plan to upgrade to 10.1, it is recommended that you make changes such that only one of these roots points to a given location. Typically, you will delete the web root before you upgrade. This eliminates redundancy and ensures that that your pipeline will display the expected files after upgrade. If you do not make a change, your files may be visible at the web root, but not at the pipeline root.
- File roots may need adjustment to make the file "Import" button appear. If you have set a pipeline root for a folder that differs from the file directory, you will not see an "Import" button in the Files web part. If possible, change project settings to use the default site-level file root instead of a pipeline root (which is set in 10.1 via a pipeline override). As a workaround, if you can't use the default root (usually because nonuniform inheritance of roots is desired), you can import files via the pipeline files web part.
- Deleted files are no longer retained. Previously, the Files web part moved deleted files into a hidden folder. As of 10.1, these files are no longer retained, so you should not rely on the hidden folder as a backup.
Lists
New list archives. You can now export all lists in a folder to an archive, then import this archive into a different folder on the same LabKey Server or a new one. This allows you to move lists efficiently.
Enhanced import process for individual lists. This includes:
- Better behavior when a user cancels an import or navigates away during import, plus a new progress bar.
- Faster canceling during import.
Assays
Assay search scope includes a shared project. You can now place an assay design in a central location such that all users in different projects can make use of it. Before 10.1, you could only place an assay design in a single project or folder and it could only be used to import assays into that particular folder. The "/Shared" project is now included in the assay search scope. It is the third namespace searched.
NAb assays. NAb now provide support for:
- Viewing graphs of different curve types. The Run Details page now allows you to choose any of of three curve fit options. See the NAb assay tutorial for an example.
- "Positive Area Under the Curve" (PAUC) calculations. The are addition to the "Area Under the Curve" (AUC) calculation added in v9.3. They appear on the Run Details page.
- Working with a broader variety of plate templates. You can now work with plates where concentration increases, not just those where it decreases. To reverse the dilution direction on the plate template, see the plate template documentation.
Enhancements to the "Run Details" page.
- You can now download all of a run’s files as a .ZIP file.
- A link to lets you jump directly to the file browser UI for a run’s directory.
Association of files with assay runs based on name. If you have uploaded files that have the same name as an assay file (but have different extensions), these related files will automatically be linked the run during the run import process. This automatic linking is useful when you have supplemental metadata files or QC reports for an assay.
Viability assay improvements. The new "Reload" button for the viability assay allows you to redo the load of a run without re-entering all of your metadata again.
Core Components
Improved grid view UI
- The enhanced Export drop-down makes it easier to export data grids to Excel (.xls and .iqy), text (.tsv) and scripts (JavaScript, R and SAS).
- Export XAR now apprears under the Export drop-down when this option is appropriate.
Better support for import/export formats.
- Multi-line and tab text are now supported during import/export of studies and lists. Special characters (tab, newline) are escaped using when exporting and unescaped on import.
Faster wiki page saves.
- This is particularly helpful for a large wiki that has a large table of contents. Save duration dropped by more than half for the labkey.org documentation tree.
Administration
Upgrades
- LabKey Server will no longer upgrade from versions of LabKey older than v2.3.
- Postgres 8.4 is now recommended, but both 8.3 and 8.4 are supported.
Password and account management
- Admin options are now available for setting strong password rules and password expiration
- The "My Account" page now provides a "Change Password" button.
- Clicking on "Log In" when already logged in will result in an immediate redirect back, which should then display the logged in status.
New Troubleshooter security role
- Troubleshooters can view site admin information but can't change it.
- Troubleshooters get an abbreviated admin menu (both in the navbar and the drop-down menu) to access the admin console.
- Most of the Diagnostics links are available.
- Troubleshooter role (and no other roles) can be assigned at the root.
Issues
- New admin option allows you to set the group of users who can be assigned an issue, along with the default assignment. You can set the "Assigned To" dropdown to show either all members of all project groups, or the members of a particular project group.
- The "Notify" field now auto-completes from the "Site Users" list.
Workbooks. A new, folder-like entity called a "workbook" provides a simple container for small-scale units of work. Key things to note:
- Workbooks do not show up in the folder tree.
- Workbooks cannot contain child folders or child workbooks (they appear more like documents than folders).
- Admins cannot set some per-folder admin options, such as set modules, missing value indicators or security. All of these things are inherited from the parent.
Study
- The dataset details view now includes a "Manage dataset" button, which allows swift admin of a dataset.
RLabKey Client API
Enhancements support the advanced R user working within a native R environment. Enhancements include:
- Improved accessibility and selection of input datasets, plus saving analysis results in a structured fashion. This includes R functions to:
- Connect to a LabKey server and set folder context
- Enumerate query objects and their fields (including lookup fields)
- Improve the flexibility of filters used in the SelectRows command
- Save data frames representing statistical results as assay data sets in LabKey
- Better control over column names. The new colNameOpt setting on selectRows and on ExecuteSql allows you to choose the way column names are determined. Previously the R client API used the caption for column names and the R report used a modified version of the field key.
JavaScript Client API
New APIs
- LABKEY.Pipeline. New class that allows programmatic manipulation of the data pipeline. Allows you to kick off certain types of pipeline jobs. Currently supports only jobs that are configured in XML, so does not support study import, export, etc.
- LABKEY.Query.getQueryDetails. Returns details about a given query including detailed information about result columns.
- LABKEY.Utils.endsWith. Determines whether one string ends with another string.
- LABKEY.ActionURL.getContainerName. Gets the current container's name instead of the container's entire path. For example, if you are in the /Project/SubFolder/MyFolder container, this method would return 'MyFolder' while getContainer() would return the entire path.
- A new "File Manager" component will be exposed in JavaScript as a beta. Changes to its APIs are expected in future releases. Developers will be able to customize the buttons displayed by this component.
API Enhancements
Java APIs
Upgrade Tip: Please recompile Java client code. The 10.1 Java API is source-compatible but not binary-compatible with 9.3. This means you will need to recompile your Java client applications using 10.1.
New Java APIs
- GetUsers. This Java Client API returns a list of users given selection criteria.
Other API Changes
New APIs available in both Java and JavaScript client libraries
- LABKEY.Security.getContainers. Returns information about the specified container, including the user's current permissions within that container. If the includeSubfolders config option is set to true, it will also return information about all descendants the user is allowed to see.
New LabKey SQL Functions and Features
- age(date1,date2). Supplies the difference in age between the two dates, calculated in years. Postgres does not support using timestampdiff on intervals larger than day. This function allows you to determine larger time differences.
- Cross-project queries. LabKey now supports cross-project queries by allowing the first item in the path to be a container path. To indicate that a query is going across projects, use a path that starts with a slash: “/Home/subfolder”.schema.query. In other words, the syntax is “/<FULL FOLDER PATH>”.<SCHEMA>.<QUERY> - for example: “/My Project/My Folder/Subfolder”.study.Participants
Schema, Table and Column Changes
Experiment (exp) Schema
- New tables for this schema. These provide details about which files and samples are used in various steps in a run.
- exp.MaterialInputs. Contains one row per material input in a protocol application for a run in this folder.
- exp.DataInputs. Contains one row per data input in a protocol application for a run in this folder.
- New columns on many Experiment (exp) tables.
- Created. Contains the date and time that this data was added to the system
- CreatedBy. Contains the user that created this data
- Modified. Contains the date and time that this data last modified
- ModifiedBy. Contains the user that last modified this data
Pipeline Schema
- New pipeline.jobs table for this schema.
- Includes data about all scheduled, running, and completed pipeline jobs.
Study Schema
- New column for study.SpecimenDetail
- ProcessingTechInitial. Identified the technician who processed the vial.
- Certain study nouns can now be customized during study creation.
- Choosing non-default nouns affects the entire study schema -- all table names and column names.
- No effect on existing tables or schemas. Only affects newly created studies.
- New StudyData table for this schema.
- Shows columns shared by datasets.
- Formerly internal-only; now exposed.
Core Schema
- New columns in the core.Containers table
- Description
- Workbook. Boolean that is set to true when the the container is a workbook.
- Title. Functions like the "Name" column, but does not need to be unique.
XML Schemas
Changes to schemas:
- study.xsd.
- timepointtype. This new attribute identifies the time measurement system used for a study. Just as before, you can use absolute visit dates or timepoints relative to a fixed start time. Additionally, as of 10.1, you can choose a timeline that supports a continuous schedule of monitoring. Terminology is still being refined as the feature is being finished. This attribute is not exposed in the UI, so it can only be used in imported studies.
- tableInfo.xsd
- propertyURI. You can now identify properties that live in a separate, shared container using either the propertyURI element or the column name of the attribute.
Improved developer menu. Developer links in the Admin menu now include a link to the
XML Schema Reference, as well as the Schema Browser and JavaScript API Reference.
Module Development
Modules can now contribute to the main credits page. To make this happen for your module, create a jars.txt file documenting all jars and drop it in the following directory: <YOUR MODULE DIRECTORY>srcMETA-INF<YOUR MODULE NAME>. The jars.txt file must be written in wiki language and contain a table with appropriate columns. See
this sample file for an example.
Modules without any Java code can now contribute pipeline configurations. Previously, modules had to include Java code to indicate that they had Spring-based pipeline configuration files available.
File-based assays can now be designed to jump to a "begin" page instead of a "runs" page. If an assay has a begin.html in the assay/<name>/views/ directory, users are directed to this page instead of the runs page when they click on the name of the assay in the assay list.
Documentation
New or enhanced sections of documentation:
- files
- fileUpload
- fileImport
- fileSharing
- adminFiles
- luceneSearch
- configDbLogin
- Comments
- Admin guide: manageComments
- User guide: comments
- Tutorial: assignComments
- Documentation for the module.properties file.
UI enhancement
- A "Help" link to the documentation now appears at the top right side of all pages of your site, next to the "My Account" link. Previously, this link appeared in the left navigation bar, which is often hidden from users.
Bug Fixes
For an exhaustive list of all issues resolved in 10.1, see
Items Completed in 10.1.
 Upgrade Tips. The enhancements mentioned above required major changes to the locations where files are stored on the file system. Tips:
Upgrade Tips. The enhancements mentioned above required major changes to the locations where files are stored on the file system. Tips: