In addition to loading and displaying the peptides and proteins identified in an MS2 run, LabKey Server lets you associate experimental annotations, which can then be pulled into the various grid views. You can display and query on things like sample properties and the experimental protocol. First, you must enter the relevant information into LabKey Server.
Loading Sample Sets
Sample sets contain a group of samples and properties for those samples. In the context of an MS2 experiment, these are generally the samples that are used as inputs to the mass spectrometer, often after they have been processed in some way.
Sample sets are scoped to a particular project inside of LabKey Server. You can reference sample sets that are in other folders under the same project, or sample sets in the "Shared" project.
To set up a sample set, first navigate to a target folder. Click on the
Experiment tab, or the
MS2 Dashboard as appropriate. By default, there will be a
Sample Sets web part. It will show all of the existing sample sets that are available in that folder. If the sample set you want to use is already loaded, select the check box in front of it and click on the
Make Active button. This will make it accessible when loading an MS2 run or for display in the grids.
If the sample set you want is not already loaded, you will need to enter the data in a tab separated format (TSV). The easiest way to do this is to use a spreadsheet like Excel. One of the columns should be the name of the sample, and the other columns should be properties of interest (the age of the participant, the type of cancer, the type of the sample, etc). Each of the columns should have a header.
Select all of the cells that comprise your sample set, including the headers, and copy them to the clipboard.
In the
Sample Sets web part, click
Import Sample Set. Give the set a useful name. Then, either cut and paste in the sample set data, or select "File" and click
Upload to select upload the spreadsheet. Click on the drop down for Id Column #1. It should contain the column headers for your sample set. Choose the column that contains the sample name or id. In most cases, you shouldn't need to enter anything for the other Id Columns. Click on
Submit. If necessary, correct any errors. One the next page, click on the
Set as Active button if it hasn't already been marked as the active sample set.
Describing mzXML files
The next step is to tie mzXML files to samples. LabKey Server will prompt you to do this when you initiate an MS2 search through the pipeline.
Go to the
Pipeline tab or the pipeline section of the MS2 dashboard and click on
Process and Upload Data. Browse to the mzXML file(s) you want to search. Click
Describe Samples.
If you've already described the mzXML files, you have the option to delete the existing information and enter the data again. This is useful if you made a mistake when entering the data the first time or want to make other changes.
If you haven't already created a protocol for your experimental procedure, click on
create a new protocol. Depending on your configuration, you may be given a list of templates from which to start. For example, you may have worked with someone at LabKey to create a custom protocol to describe a particular fractionation approach. Select a template, if needed, and fill in a description of the protocol.
Then select the relevant protocol from the list. If you started from a directory that contains multiple mzXML files, you will need to indicate if the mzXML files represent fractions of a larger sample.
The next screen asks you to identify the samples that were inputs to the mass spectrometer. The active sample set for the current LabKey Server folder, if any, is selected as the default sample set. It is strongly recommended that you use the active sample set or no sample set. You can change the default name for the runs. For each run, you are asked for the Material Sample ID. You can use the text box to type in a name if it is not part of a sample set. Otherwise, choose the name of the sample from the drop down.
Once you click on
Submit, LabKey Server will create a XAR file that includes the information you entered and load it in the background.
Kicking off an MS2 search
To initiate an MS2 search, return to the Data Pipeline and browse back to the mzXML files. This is described in the
Search and Process MS2/MS2 Data topic.
Viewing Annotation Data
There are a number of different places you can view the sample data that you associated with your mzXML files. First, it's helpful to understand a little about how LabKey Server stores your experimental annotations.
A set of experimental annotations relating to a particular file or sample is stored as an
experiment run. Each experiment run has a
protocol, which describes the steps involved in the experimental procedure. For MS2, LabKey Server create an experiment run that describes going from a sample to one or more mzXML files. Each time you do a search using the mzXML files creates another experiment run. LabKey Server can tie the two types of runs because it knows that the output of the first run, the mzXML files, are the inputs to the search run.
You can see the sample data associated with a search run using the Enhanced MS2 Run view, or by selecting the "MS2 Searches" filter in the Experiment tab's run list. This view will only show MS2 runs that have experimental data loaded. In some cases, such as if you moved MS2 runs from another folder using LabKey Server 1.7 or earlier, or if you directly loaded a pep.xml file, no experimental data will be loaded.
Click on the
Customize View button. This brings up the column picker for the run list. Click to expand the
Input node. This shows all the things that might be inputs to a run in the current folder. Click to expand the
mzXML node. This shows data for the mzXML file that was an input to the search run. Click to expand the
Run node. This shows the data that's available for the experiment run that produced the mzXML file. Click to expand the
Input node. This shows the things that might be inputs to the run that produced the mzXML file. If you used a custom template to describe your mass spectrometer configuration, you should expand the node that corresponds to that protocol's inputs. Otherwise, click to expand the
Material node. Click to expand the
Property node, which will show the properties from the folder's active sample set. Click to add the columns of interest, and then
Save the column list.
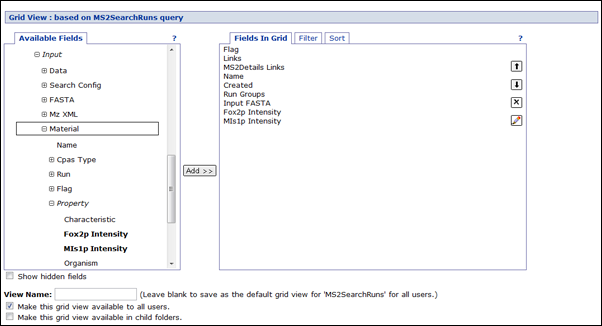
You can then filter and sort on sample properties in the run.
You can also pull in sample information in the peptides/proteins grids by using the new Query grouping. Use the column picker to go to Fraction->Run->Experiment Run. At this point, you can follow the instructions above to chain through the inputs and get to sample properties.
 You can then filter and sort on sample properties in the run.You can also pull in sample information in the peptides/proteins grids by using the new Query grouping. Use the column picker to go to Fraction->Run->Experiment Run. At this point, you can follow the instructions above to chain through the inputs and get to sample properties.
You can then filter and sort on sample properties in the run.You can also pull in sample information in the peptides/proteins grids by using the new Query grouping. Use the column picker to go to Fraction->Run->Experiment Run. At this point, you can follow the instructions above to chain through the inputs and get to sample properties.