A key component of using Luminex® instrument data is calculation of logistic curve fits as well as other values for standards, unknowns, and quality controls. During the second Luminex tutorial, in
Step 4: View 4pl and 5pl Curve Fits, you saw the 4pl and 5pl logistic curve fits, and then used some of the calculated values in
Step 5: Track Analyte Quality Over Time.
Some calculations are done by the BioPlex software itself, LabKey Server performs others, and still more are made using R and the Ruminex package by way of the LabKey Luminex transform script. By further customizing the assay design and adding additional operations to the transform script, additional calculations may be added enabling you to tailor the instrument data framework to suit your specific research needs.
Background
LabKey Luminex Transform Script Calculations
The LabKey Luminex transform script uses the Ruminex R package to calculate logistic curve fits for each titration for each analyte. Titrations may be used either as standards or as quality controls. In this tutorial, all titrations are used as standards.
Curve fits are calculated using both a 4 parameter logistic (4pl) and a 5 parameter logistic (5pl). Based on these curve fits, the script calculates
EC50s ("Expected Concentration at %50") for the standard for each analyte. An EC50 is the concentration or dilution that is expected to produce
half of the difference between the asymptotic maximum and minimum fluorescence intensities.
For each run, the script outputs a PDF that includes plots for curve fits for each analtye. Each plot shows the dose response curve for fluorescence intensity with increasing concentration or reduced dilution. These plots can be useful for examining how well the curves fit the data.
LabKey Server Calculations
LabKey Server itself calculates the
AUC ("Area Under the Curve") for each standard titration using a trapezoidal calculation based on observed values. LabKey Server also identifies the
HighMFI ("Highest Mean Fluorescence Intensity") for each titration.
BioPlex Calculations vs. LabKey Luminex Transform Script Calculations
The
Excel run files produced by the BioPlex instrument also include 5-parameter logistic curve fits for each titrated standard and each titrated quality control. These 5pl regressions were calculated by the BioPlex software. The Excel run files also include estimates for sample concentrations generated using each sample's measured
FI-Bkgd (fluorescence intensity minus background well fluorescence intensity) and the 5pl regression equations derived for the standards by the instrument software. LabKey Server reports these results alongside all other imported run data, as shown in the
Obs Conc BioPlex 5PL column in the default
Results view for any run (as shown
here for run 10).
However, these instrument-generated 5pl regressions and estimates for sample concentrations are not the same as the results given by the LabKey Luminex transform script. The script's results for the parameters of the 5pl regression (
Slope Param 5 PL, Lower Param 5 PL, Upper Param 5 PL, Inflection Param 5 PL, and
Asymmetry Param 5 PL) and the estimated concentrations of samples (
Est Conc - Rumi 5 PL) are reported as columns in the
Results view for any run.
Using the script for 5pl regression calculations allows subtraction of negative bead fluorescence intensity before fits and other customizations, such as the treatment of negative values, weighting of standard errors, and optional log transforms of data. Users may further customize how the script performs 5pl curve fits as they see fit.
Only the 5pl results calculated by the script, not the BioPlex instrument software, are used by LabKey Server. For example, only script-calculated 5pl EC50s are shown in Levey-Jennings plots. All references to 5pl curve fits or EC50s in this tutorial refer to fits and results calculated by the script, not the instrument software, unless specifically noted otherwise.
Review Calculated Values
In
Step 4: View 4pl and 5pl Curve Fits you reviewed calculated values as they are available in both titration qc reports or in the results grid for the run data. The titration qc report includes summary results for both
standards and
QC controls for a given run or for all runs. The results grid includes regression parameters for all curve fits.
Subtract Negative Control Bead Values
The
FI-Bkgd-Neg column shows the fluorescence intensity after both the background well and the negative bead are subtracted. The assay design used in the
Luminex Assay Tutorial Level II tells the script (via the
StandardCurveFitInput property's default value) to use fluorescence alone (
FI), without subtraction, to fit 4pl and 5pl curves to titrated
standards. In contrast, the assay design tells the script (via the
UnknownFitInput property's default value) to use
FI-Bkgd-Neg to estimate concentrations for
unknowns using the 4pl and 5pl regression equations calculated from the standards.
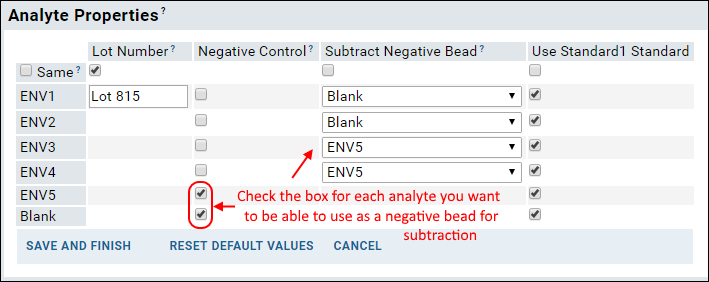
When calculating the value FI-Bkgd-Negative (fluorescence intensity minus background FI minus a negative bead), you may specify on a per analyte basis what to use as the negative bead. Depending on the study, the negative bead might be blank, or might be a more suitable negative control antigen. For example, in a study of certain HIV antigens, you might subtract MulV gp70 from gp70V1V2 proteins, blank from other antigens, etc. Note that the blank bead is not subtracted by default - it must be explicitly selected like any other negative bead.
To enable subtraction of negative control bead values, the assay design must be modified to include a run field of type Boolean named
NegativeControl. Then during the assay run import, to select negative beads per analyte, you'll set
Analyte Properties when importing data files. First identify specific analytes using checkboxes in the
Negative Control column, then select one of these negative beads for each analyte in the
Subtract Negative Bead column.
Skip Ruminex Calculations
If you want to have the option to skip the Ruminex calculations of estimated concentration values and generation of standard curve fit PDFs, you can do so by adding a
SkipRumiCalculation property to the assay design. All 4PL EC50 and AUC values are calculated regardless of the flag, but when it is defined the user will have the option to check a box on import to skip those calculations.
To explore this optional feature, use a copy of the assay design. The tutorial uses the original design and may not work as expected if you skip the Ruminex calculations.
Note: You may need to replace the transform script included with the tutorial with one that knows about the SkipRumiCalculation property. Click to download this file to replace the version you copied to your "scripts" directory
when you began the tutorial:
This version of the script will still work correctly with assay designs which do not use this property.
- From the Assay Dashboard, click Luminex Assay 200.
- Select Manage Assay Design > copy assay design.
- Click Copy to Current Folder.
- Name the new copy "Luminex Skip Rumi 200".
- In Run Fields, click Add Field and add:
- Name: SkipRumiCalculation (no spaces)
- Type: Boolean

Your new assay design has been added to the list and can be used to import an example run:
- Click Luminex Skip Rumi 200.
- Click Import Data.
- Click Next, then in Run Properties:
- Enter an Assay Id, such as "Run2skip" (since you will be uploading a file you already imported, this name must be unique).
- Check the box for Skip Rumi Calculation, which you just added by defining the property.
- In the Run Data field, click Browse or Choose File
- Select a run from the demo files on your local machine, for example: /Luminex/Runs - Assay 200/02-14A22-IgA-Biotin.xls
- Click Next then Save & Finish.
Notice that the standard curve PDFs were not created and if you click the Assay ID
Run2skip and scroll right, the various *Rumi* estimated concentration values have not been calculated.
Use Uploaded Positivity Threshold Defaults
If your lab uses specific positivity cutoff values, you can manually enter them on an antigen-by-antigen basis during upload on the
Analyte Properties panel. To simplify user entry and reduce the possibilities of errors during this process, you may specify analyte-specific default values for the
PositivityThreshold property on a per folder and assay design combination. The default default value is 100. To specify analyte-specific defaults, add them to the assay design for a given folder as described here using the
Luminex Assay Tutorial Level II example:
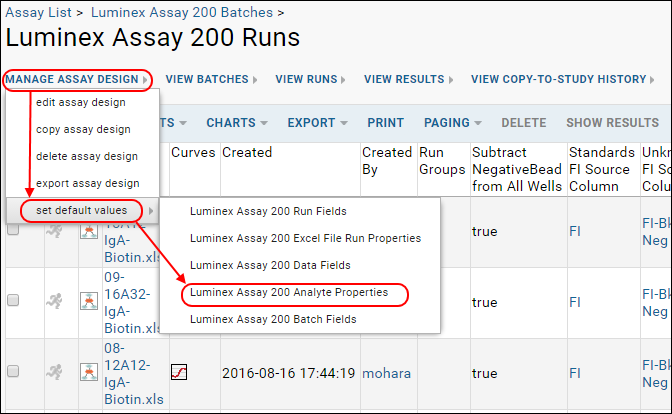
- From the Assay Dashboard, click Luminex Assay 200.
- Select manage assay design > set default values > Luminex Assay 200 Analyte Properties.
- Enter Analyte and desired Positivity Threshold.
- Click the + button to add another.
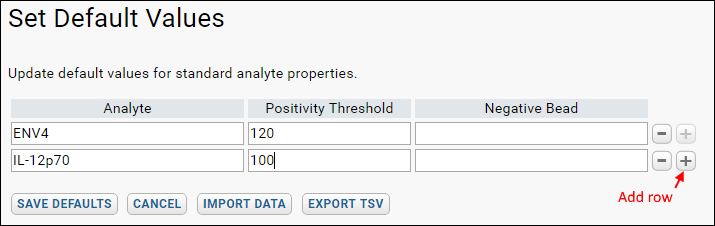
- You may instead click Import Data to upload a TSV file or copy and paste the data to simplify data entry.
- Click Save Defaults when finished.
When you import a set of positivity threshold data, it overwrites the prior set, meaning that any defaults previously defined but missing from the imported TSV will be dropped.

