When features are not in widespread use, but can be made available if needed, they will be available on one of these pages on the
> Site > Admin Console under
Configuration:
- Optional Features: Optional features are not typically used; discuss with your Account Manager before enabling any optional feature.
- Deprecated Features: Features that have been deprecated, but administrators may need to temporarily enable in order to migrate customizations to supported features.
- Experimental Features: Features under development or otherwise not ready for production use.
Note that some features listed here are only available when specific modules are included. Proceed with discretion and
please contact LabKey if you are interested in sponsoring further development of anything listed here. Enabling or disabling some features will require a restart of the server.
Optional Features
Optional features are not typically used; discuss with your account manager before enabling any optional feature.
Stage file uploads and downloads to temporary local file
When using a non-local file system, using a specific API that requires a locally staged copy of the file as the source can sometimes be significantly faster than streaming the file directly to/from storage
Stage SKYD files to a temporary local file for import purposes
When using a non-local file system, the latency for random access requests can be significantly slower than first copying to local storage.
Deprecated Features
WARNING: Deprecated features will be removed very soon, most likely for the next major release. If you enable one of these features you should also create a plan to stop relying on it.
Restore 'Advanced Import Options' during Folder import
The options to select specific folder objects during folder archive import and to have an import applied to multiple folders will be removed in LabKey Server v25.7.
Restore Object-Level Discussions
This option and all support for Object-Level Discussions will be removed in LabKey Server v25.7.
Study Protocol Design Tools
This option adds support for the study protocol and vaccine design tools.
Experimental
WARNING: Experimental features may change, break, or disappear at any time. We make absolutely no guarantee about what will happen if you turn on any experimental feature.
- Select > Site > Admin Console
- Under Configuration, click Experimental Features.
- Carefully read the warnings and descriptions before enabling any features. Searching the documentation for more detail is also recommended, though not all experimental features are documented.
- Use checkboxes to enable desired experimental features. Uncheck the box to disable the feature.
Allow gap with withCounter and rootSampleCount expression
Check this option if gaps in the count generated by withCounter or rootSampleCount name expression are allowed. Changing this setting requires a server restart.
Allow merge of study dataset that uses server-managed additional key fields
Merging of dataset that uses server-managed third key (such as GUID or auto RowId) is not officially supported. Unexpected outcomes might be experienced when merge is performed using this option.
Allow query based dataset snapshots
Allow unprovisioned, query-based dataset snapshots to be created.
Assay linked to study consistency check
Flags rows in assay-linked datasets where the subject and timepoint may be different from the source assay. When enabled, if rows in the linked study dataset are not consistent with the source assay, you'll see them highlighted. For example, if an assay result row was edited after being linked (in this case to change the visit ID), the dataset would alert the user that there was something to investigate:
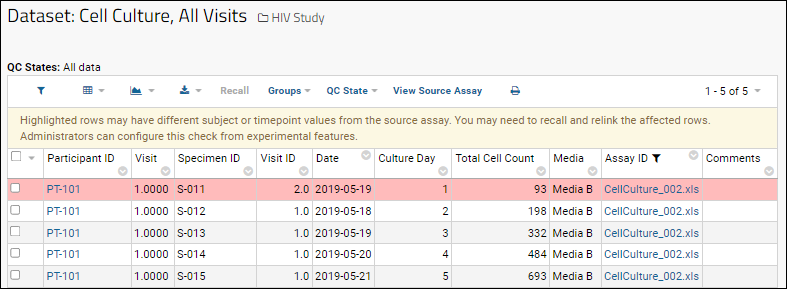
Recalling and relinking the affected rows should clear the inconsistency.
Block Malicious Clients
Reject requests from clients that appear malicious. Make sure this feature is disabled before running a security scanner.
Data Region Async Total Rows
Enable asynchronous calculation of total rows for data regions. This can improve performance for large datasets.
Disable enforce Content Security Policy
Stop sending the Content-Security-Policy header to browsers, but continue sending the Content-Security-Policy-Report-Only header. This turns off an important layer of security for the entire site, so use it as a last resort only on a temporary basis (e.g., if an enforce CSP breaks critical functionality).
Disable Login X-FRAME-OPTIONS=DENY
By default LabKey disables all framing of login related actions. Disabling this feature will revert to using the standard site settings.
Disable SQLFragment strict checks
SQLFragment now has very strict usage validation, these checks may cause errors in code that has not been updated. Turn on this feature to disable checks.
Generic [details] link in grids/queries
This feature will turn on generating a generic [details] link in most grids.
Include Last-Modified header on query metadata requests
For schema, query, and view metadata requests, enabling this will include a Last-Modified header such that the browser can cache the response. The metadata is invalidated when performing actions such as creating a new list or modifying the columns on a custom view.
Less restrictive product folder lookups
Allow for lookup fields in product folders to query across all containers within the top-level project.
Media data folder export and import
Enable a subset of Media data for folder export and processing of data for folder import. This does not provide a full-fidelity transfer of data.
Media Ingredient read permissions
Enforce media ingredient read permissions.
No Guest Account
When this experimental feature is enabled, there will be no guest account. All users will have to create accounts in order to see any server content.
Notebook Custom Fields
Enable custom fields in Electronic Lab Notebooks. Available in Biologics LIMS only. Learn more in this topic:
Notifications Menu
Display a notifications 'inbox' icon in the header bar with a display of the number of notifications; click to show the notifications panel of unread notifications.
Resolve Property URIs as Columns on Experiment Tables
If a column is not found on an experiment table, attempt to resolve the column name as a Property URI and add it as a property column.
Sample/Aliquot Selector
Enable the Sample/Aliquot Selector button to show in sample grids within Sample Manager and Biologics.
Use QuerySelect for row insert/update form
This feature will switch the query based select inputs on the row insert/update form to use the React QuerySelect component. This will allow for a user to view the first 100 options in the select but then use typeahead search to find the other select values.
Use row-by-row update
For Query.updateRows api, do row-by-row update, instead of using a prepared statement that updates rows in batches.
Use Sub-schemas in Study
Separate study tables into three groups: Datasets, Design, Specimens. User defined queries are not placed in any of these groups.
