File Watchers let administrators
set up the monitoring of directories on the file system and perform specific actions when desired files appear. This topic describes the various pipeline tasks available. The configuration options for File Watcher triggers and specifics about type of files eligible vary based on the type of File Watcher, and the tasks available vary by folder type.
File Watcher Tasks
Reload Folder Archive
This option is available in any folder type and reloads an unzipped folder archive. It will not accept compressed (.zip) folder archives.
The reloader expects a folder.xml in the base directory of the archive. To create an unzipped folder archive,
export the folder to your browser as a .zip file and then unzip it.
To reload a study, use this File Watcher, providing an unzipped folder archive containing the study objects as well as the folder.xml file and any other folder objects needed.
Import Samples From Data File
This option supports importing Sample data into a specified Sample Type. A few key options on the
Configuration panel are described here. You may also want to set the
auditBehavior custom parameter to control the level of audit logging.
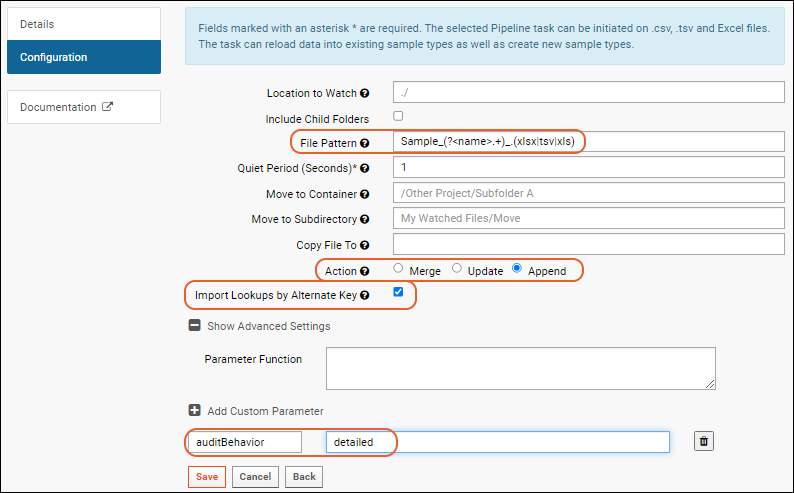
File Pattern
You can tell the trigger which Sample Type the imported data belongs to by using one of these
file name capture methods:
- <name>: the text name of the Sample Type, for example "BloodVials".
- <id>: The integer system id of the Sample Type, for example, "330". To find the system id: go to the Sample Types web part and click a Sample Type. The URL will show the id as a parameter named 'RowId'. For example:
https://SERVER_NAME/FolderPath/experiment-showSampleType.view?rowId=330
For example, a
File Pattern using the
name might look like:
Sample_(?<name>.+)_.(xlsx|tsv|xls)
...which would recognize the following file name as targeting a Sample Type named "BloodVials":
If the target Sample Type does not exist, the File Watcher import will fail.
Action: Merge, Update, or Append
Import behavior into Sample Types has three options:
- Merge: Update existing samples as described below and insert new samples from the same source file.
- Update: Provided all incoming rows match existing rows, update the data for these rows.
- When an incoming field contains a value, the corresponding value in the Sample Type will be updated. When a field in the imported data has no value (an empty cell), the corresponding value in the Sample Type will be deleted.
- If any rows do not match existing rows, the update will fail.
- Append: The incoming data file will be inserted as new rows in the Sample Type. The operation will fail if there are existing sample ids that match those being imported.
Import Lookups by Alternate Key
Some sample fields, including the built in
Status field, are structured as lookups. When a File Watcher encounters a value other than the primary key for such a lookup, it will only resolve if you check the box to
Import Lookups by Alternate Key.
For example, if you see an error about the inability to convert the "Available (String)" value, you can either:
- Edit your spreadsheet to provide the rowID for each "Status" value, OR
- Edit your File Watcher to check the Import Lookups by Alternate Key box.
Import Assay Data from a File
Only Standard assay designs are supported, under the
General assay provider. Multi-run files and run re-imports are not supported by the File Watcher.
When you create this file watcher, select the
Assay Provider "General", meaning the Standard type of assays.
The following file formats are supported, note that .txt files are not supported:
- xls, .xlsx, .csv, .tsv, .zip
The following assay data and metadata are supported by the File Watcher:
- result data
- batch properties/metadata
- run properties/metadata
If only result data is being imported, you can use a single tabular file.
If additional run metadata is being imported, you can use either a zip file format or an excel multi-sheet format. In a zip file format the system determines the data type (result, run metadata, etc.) using the names of the files. In the multi-sheet format the system matches based on the sheet names. The sheet names don't need to be in any particular order. The following matching criteria are used:
| data type | for zipped files, use file name... | for multi-sheet Excel, use sheet name... |
|---|
| batch properties | batchProperties.(tsv, csv, xlsx, xls) | batchProperties |
| run properties | runProperties.(tsv, csv, xlsx, xls) | runProperties |
| results data | results.(tsv, csv, xlsx, xls) | results |
The following multi-sheet Excel file shows how to format results data and run properties fields on different sheets:
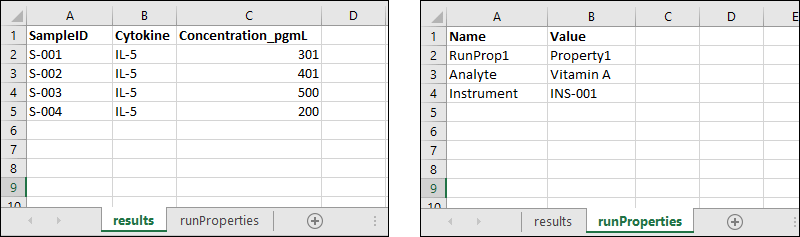
When run properties metadata is included, either via a file in the .zip or a spreadsheet tab, there is special handling to "extract" the runProperties first, so that it can then be handed off to the usual assay import process. If, for example, any transform scripts are included with the assay, they will be run with the run properties available as usual.
Configure Target Assay Design
The assay provider (only General is supported) and protocol (assay design name) can be specified in the File Watcher configuration. This is easier to configure than binding to the protocol using a regular expression named capture group.
If there is no name capture group in the file pattern and there is a single assay protocol in the container, the system attempts to import into that single assay. If the target assay does not exist or cannot be determined, the File Watcher import will fail.
Use Name Capture to Target Any Assay Design
When setting the
File Pattern,
regular expression 'name capture' can be used as with other File Watcher types to match names or IDs from the source file name.
Two capture groups can be used:
- name: the assay protocol name (for example, MyAssay)
- id: the system id of the target assay (an integer)
For example this file name pattern:
assay_(?<name>.+)_.(xlsx|tsv|xls|zip)
will interpret the following file name as targeting an assay named "MyAssay":
The following example file pattern uses the protocol ID instead of the assay name:
assayProtocol_(?<id>.+)_.(xlsx|tsv|xls|zip)
which will interpret this file as targeting the assay with protocol ID 308:
Reload Lists Using Data File
This option is available in any folder type, provided the list module has been enabled. It imports data to existing lists from source files in either Excel (.xls/.xlsx) or TSV (.tsv) formats. It can also infer non-key column changes. Note that this task cannot create a new list definition: the list definition must already exist on the server.
You can reload lists from files in S3 storage by enabling an SQS Queue and configuring cloud storage to use in your local folder. Learn more in this topic:
Move Files Across the Server
This option is available in any folder type. It moves and/or copies files around the server without analyzing the contents of those files.
Import Study Data from a CDISC ODM XML File
This option is provided for importing electronically collected data in the CDISC ODM XML format (such as from tools like
DFdiscover). It is available only when the
CDISC_ODM module is enabled in a given folder.
Learn more in this topic:
Import/Reload Study Datasets Using Data File
This option is available in a study folder. It loads data into existing study datasets and it infers/creates datasets if they don't already exist. You can configure the File Watcher to either:
- Append: Add new data to the existing dataset
- Replace: Replace existing data with the new data
The following file formats are supported, note that .csv files are not supported:
- .tsv, .txt, xls, .xlsx, .zip
You can use a
name capture group to be able to identify the target dataset as a portion of the filename. You can also use a compound name capture group to have a File Watcher target multiple studies from the same location. Examples are available in this topic:
File Watcher: File Name PatternsIf you don't use a name capture group, the system will use the entire filename stem as the name of the dataset. For example, dropping the following files into the watched location will load two datasets of these names:
| Dropped File | Dataset Loaded |
|---|
| Demographics.xls | Demographics |
| LabResults.xls | LabResults |
| New_LabResults.xls | New_LabResults |
To have a file like "New_LabResults.xls" reload new data into the LabResults dataset, you would need a name capture group that parsed out the <name> between the underscore and dot.
Import Specimen Data Using Data File
This option is only available in study folders with the specimen module enabled.
This File Watcher type accepts specimen data in both .zip and .tsv file formats:
- .zip: The specimen archive zip file has a .specimens file extension.
- .tsv: An individual specimens.tsv file which will typically be the simple specimen format and contain only vial information. This file will have a # specimens comment at the top.
By default, specimen data imported using the a File Watcher will be set to
replace existing data. To
merge instead, set the
custom property "mergeSpecimen" to true.
Specimen module docs:
Specimen Tracking (Legacy)
Import a Directory of FCS Files (Flow File Watcher)
Import flow files to the flow module. This type of File Watcher is only available in Flow folders. It supports a process where FCS flow data is deposited in a common location by a number of users. It is important to note that each data export must be placed into a new separate subdirectory of the watched folder. Once a subfolder has been 'processed', adding new files to it will not trigger a flow import.
When the File Watcher finds a new subdirectory of FCS files, they can be placed into a new location under the folder pipeline root based on the current user and date. Example: @pipeline/${username}/${date('YYYY-MM')}. LabKey then imports the FCS data to that container. All FCS files within a single directory are imported as a single experiment run in the flow module.
Quiet Period for Flow File Watchers
One key attribute of a flow File Watcher is to ensure that you set a long enough
Quiet Period. When the folder is first created, the File Watcher will "wait" the specified quiet period before processing files. This interval must be long enough for all of the files to be uploaded, otherwise the File Watcher will only import the files that exist at the end of the quiet period. For example, if you set a 1 minute quiet period, but have an 18 file FCS folder (such as in our tutorial example) you might only have 14 files uploaded at the end of the minute, so only those 14 will be imported into the run. When defining a flow File Watcher, be sure to set an adequate quiet period. In situations where uploads take considerable time, you may decide to keep using a manual upload and import process to avoid the possibility of incomplete runs.
In addition, if your workflow involves creating subfolders of files, the creation of each new subfolder will trigger a new quiet period delay, which can lead to the perception of multiplied wait times.
Custom File Watcher Tasks
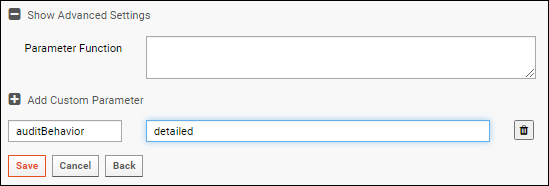
Custom Parameters
Add custom parameters on the
Configuration panel, first expanding the
Show Advanced Settings section. Click
Add Custom Parameter to add each one. Click
to delete a parameter.

allowDomainUpdates
This parameter used in earlier versions has been replaced with the checkbox option to
Allow Domain Updates on the
Configuration panel for the tasks 'Reload Lists Using Data File' and 'Import/Reload Study Datasets Using Data File'.
When updating lists and datasets, by default, the columns in the incoming data will overwrite the columns in the existing list or dataset. This means that any new columns in the incoming data will be added to the list and any columns missing from the incoming data will be dropped (and their data deleted).
To override this behavior, uncheck the
Allow Domain Updates box to retain the column set of the existing list or dataset.
default.action
The "default.action" parameter accepts text values of either : replace or append, the default is replace. This parameter can be used to control the default
Action for the trigger, which may also be
more conveniently set using the Action options on the
Configuration panel.
mergeData
This parameter can be included to merge data, with the value set to either true or false. The default is false (replace) and for existing configurations if no param was provided we interpret that as : false/replace.
Where supported, merging can be
more conveniently set using the Action options on the
Configuration panel.
mergeSpecimen
By default, specimen data imported using the 'Import Specimen Data Using Data File' File Watcher will be set to
replace existing data. To
merge instead, set the property "mergeSpecimen" to true.
skipQueryValidation
'Reload Study' and 'Reload Folder Archive' can be configured to skip query validation by adding a custom parameter to the File Watcher named 'skipQueryValidation' and setting it to 'TRUE'. This may be helpful if your File Watcher reloads are failing due to unrelated query issues.

auditBehavior
The 'Import Samples from Data File' task supports the 'auditBehavior' custom parameter to control the level of detail that will be logged. Valid options are:
If your File Watcher will be loading sample data into either the Sample Manager or Biologics LIMS products, it is suggested that you set this parameter to "detailed".
Related Topics
 When run properties metadata is included, either via a file in the .zip or a spreadsheet tab, there is special handling to "extract" the runProperties first, so that it can then be handed off to the usual assay import process. If, for example, any transform scripts are included with the assay, they will be run with the run properties available as usual.
When run properties metadata is included, either via a file in the .zip or a spreadsheet tab, there is special handling to "extract" the runProperties first, so that it can then be handed off to the usual assay import process. If, for example, any transform scripts are included with the assay, they will be run with the run properties available as usual.
