The protein details page can be reached by clicking the name/label from the Precursor list. Depending on the contents of the Skyline file, you will some or all of the features described in this topic. Users have the option to see multiple peptides or small molecules in the same chromatogram.
After loading your Skyline document into Panorama, click the name of the file to open the details page. Then click the name of a protein / label to open the
Protein/Molecule List Details page.
Precursor List
The
Precursor List shows summary information about the Peptides and Proteins contained in the Skyline file. Use the
and to expand/collapse the Peptide section. Click the name in the first text column to see
more detail.
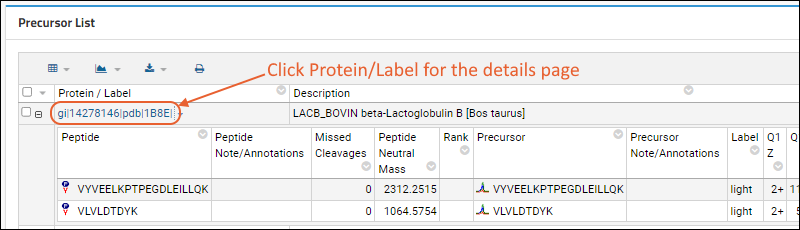
Hover over the
next to the name for a tooltip with more details about the sequence.

Protein Highlights
The top section of any single protein detail page shows various highlights, depending on the details present in the Skyline file.
- Name: The Skyline document's file name as a link, plus a menu of download options.
- Description: If any.
- Decoy: Whether the protein is marked as decoy.
- Accession: An Accession link if present.
- Note: If any.
Protein Grouping
If the Skyline file includes
Protein Grouping, in which one or more peptides match not to a single protein sequence, but multiple proteins, the precursor list will show the group of proteins. The protein group label is typically the concatenated and slash-separated list of protein names, but this is not required.
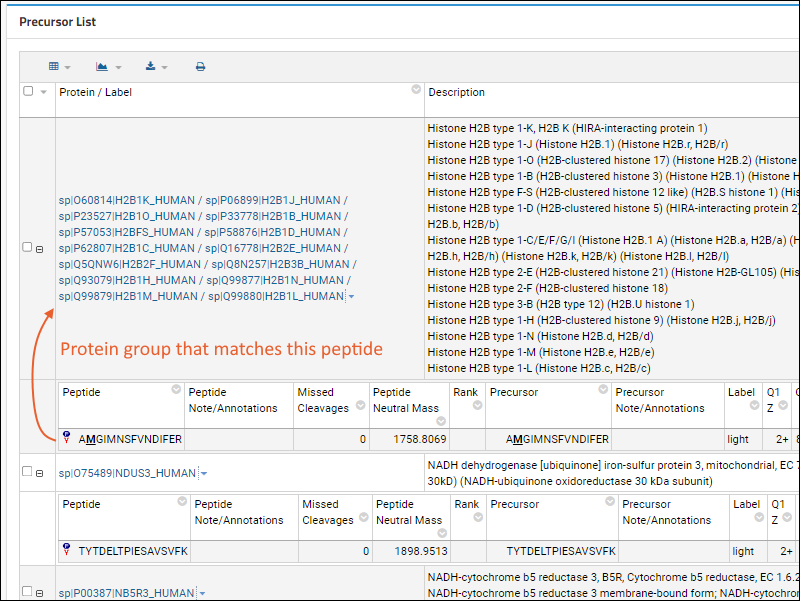
When you click the name of a protein group, the details page will show a
highlights section for the group, then list all of the proteins in a grid. If there are protein sequences available, Panorama will automatically show the first protein's sequence (as for a standalone protein). Users can click on any of the other protein names to see the details page for that sequence.
Sequence Coverage
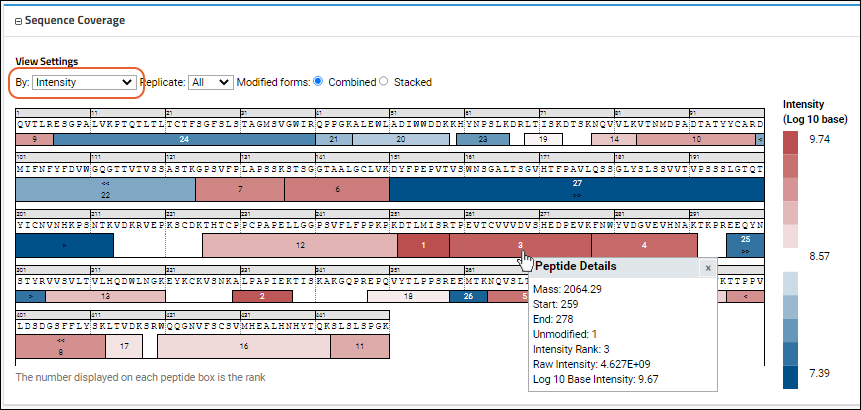
The
Sequence Coverage map provides a heatmap style view giving the user an immediate visual assessment of:
- Peptide XIC intensity
- Peptide ID confidence data
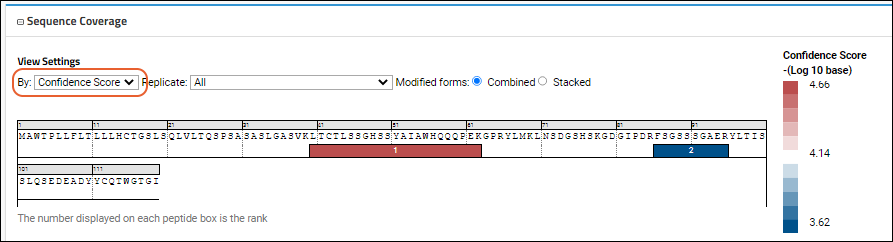
On the map, color correlates intensity or confidence score, with the rank displayed on each peptide box in both views. The map helps you visualize the spatial location within the sequence. The legend maps colors to the values they represent. Hovering over any peptide box will give more details, including a grid of multiple modified forms of the peptide if they exist. The
Intensity view might look like this:
Use the
View Settings to control the display:
- By:
- Intensity (shown above): Use the Log 10 based intensity value
- Confidence Score (shown below)
- Replicate:
- All: (Default) Displays the maximum intensity or confidence of the peptide across all replicates.
- Individual replicates present
- Modified forms: In cases where there is more than one modified version of a peptide (modified/unmodified, multiple modifications), select how to display them:
- Combined: Use the sum of intensities of all the modified forms and show in a single box.
- Stacked: Show a 'stacked' set of boxes for each of the modified forms, with the modification with the highest intensity shown on top.
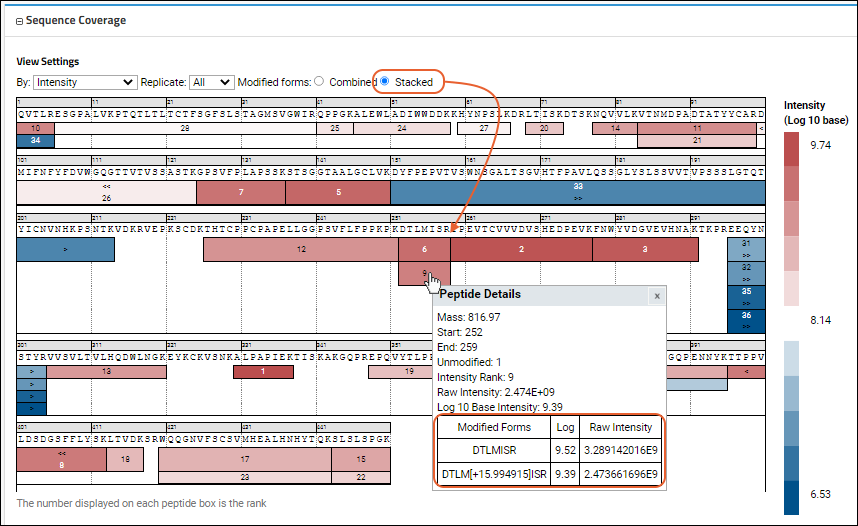
The
Confidence Score view of another sequence might look like this:
When there is no intensity data, or no confidence scores are present in the file, there will be a 'non-heatmap' version shown. For example:

Multiple Peptides Per Chromatogram
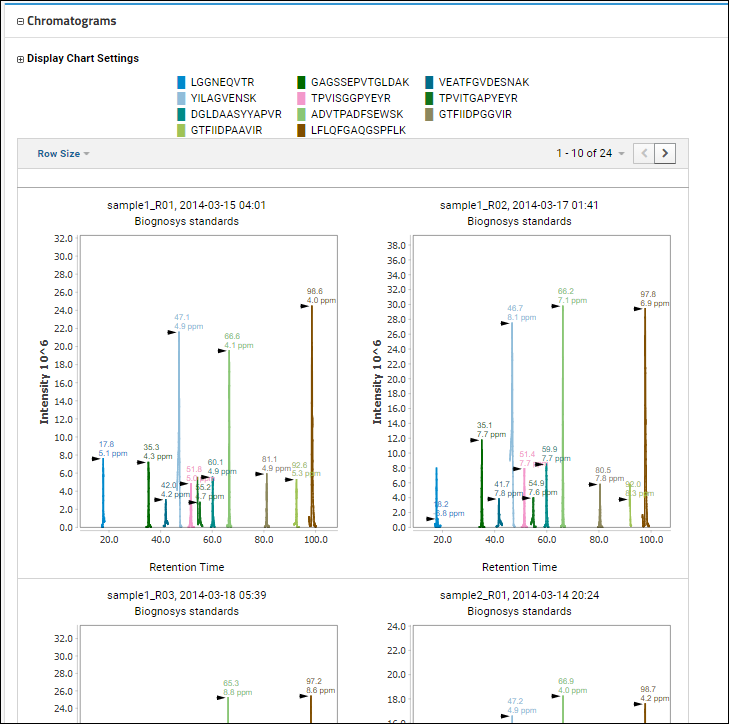
When the protein contains many peptides, it can be useful to see the chromatograms for several peptides in one plot.
On the protein details page, scroll down to the
Chromatograms section where you will a legend showing the colors assigned to the individual peptides present. By default all peptides are included, unless you
select a subset using the checkboxes. The chromatograms below all use this scheme to show the same series of peptides for each replicate.
Use the
Row Size menu to control the display size of the SVG plots; options are 1, 2, 3, 4, 5, or 10 per row.
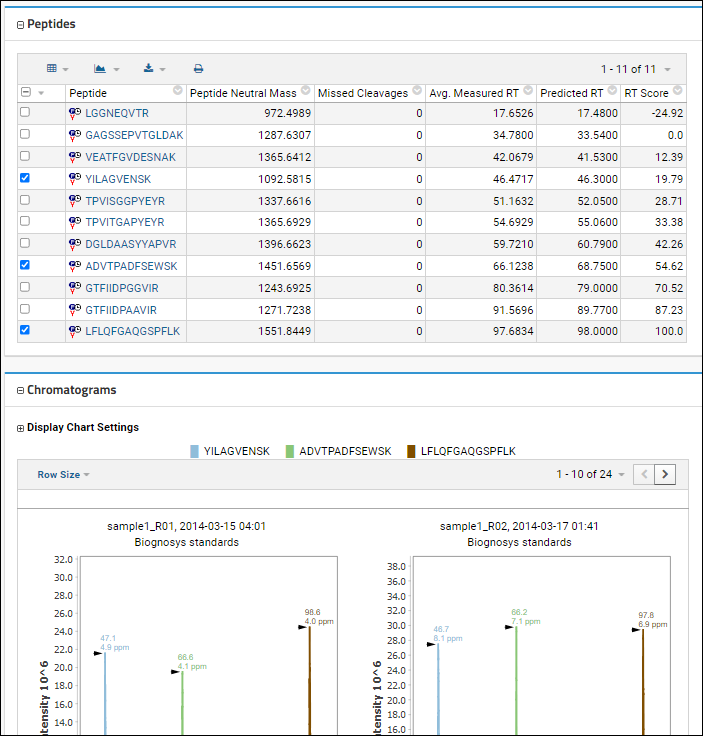
Select Peptides for Chromatograms
By default, all peptides will be included in the chromatograms, unless you use the checkboxes in the
Peptides panel to select only a subset.
You can also use the
Display Chart Settings to to multi-select the specific replicates for which you want to see the chromatograms, making it easier to compare a smaller number of interest.
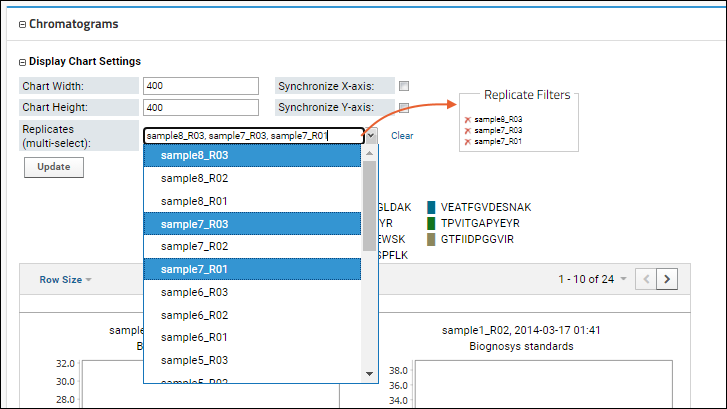
Learn more about Chromatogram Display Settings in this topic:
Related Topics