The
script pipeline lets you run scripts and commands in a sequence, where the output of one script becomes the input for the next in the series. Automating data processing using the pipeline lets you:
- Simplify procedures and reduce errors
- Standardize and reproduce analyses
- Track inputs, script versions, and outputs
The pipeline supports running scripts in the following languages:
Note that JavaScript is
not currently supported as a pipeline task language. (But pipelines can be
invoked by external JavaScript clients, see below for details.)
Pipeline
jobs are defined as a sequence of
tasks, run in a specified order. Parameters can be passed using
substitution syntax. For example a job might include three tasks:
- pass raw data file to an R script for initial processing,
- process the results with Perl, and
- insert into an assay database.
Topics
Premium Feature AvailablePremium edition subscribers can configure a
file watcher to automatically invoke a properly configured script pipeline when desired files appear in a watched location. Learn more in this topic:
Set Up
Before you use the script pipeline, confirm that LabKey is configured to use your target script engine.
You will also need to deploy the module that defines your script pipeline to your LabKey Server, then
enable it in any folder where you want to be able to invoke it.
Module File Layout
The module directory layout for sequence configuration files (.pipeline.xml), task configuration files (.task.xml), and script files (.r, .pl, etc.) has the following shape. (Note: the layout below follows the pattern for modules as checked into LabKey Server source control. Modules not checked into source control have a somewhat different directory pattern. For details see
Map of Module Files.)
<module>
resources
pipeline
pipelines
job1.pipeline.xml
job2.pipeline.xml
job3.pipeline.xml
...
tasks
RScript.task.xml
RScript.r
PerlScript.task.xml
PerlScript.pl
...
Parameters and Properties
Parameters, inputs, and outputs can be explicitly declared in the xml for tasks and jobs, or you can use substitution syntax, with dollar-sign/curly braces delimeters to pass parameters.
Explicit definition of parameters:
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ScriptTaskType"
name="someTask" version="0.0">
<inputs>
<file name="input.txt" required="true"/>
<text name="param1" required="true"/>
</inputs>
Using substitution syntax:
<task xmlns="http://labkey.org/pipeline/xml" name="mytask" version="1.0">
<exec>
bullseye -s my.spectra -q ${q}
-o ${output.cms2} ${input.hk}
</exec>
</task>
Substitution Syntax
The following substitution syntax is supported in the script pipeline.
| Script Syntax | Description | Example Value |
|---|
| ${OriginalSourcePath} | Full path to the source | "/Full_path_here/@files/./file.txt" |
| ${baseServerURL} | Base server URL | "http://localhost:8080" |
| ${containerPath} | LabKey container path | "/PipelineTestProject/folderName" |
| ${apikey} | A temporary API Key associated with the pipeline user | "3ba879ad8068932f4699a0310560f921495b71e91ae5133d722ac0a05c97c3ed" |
| ${input.txt} | The file extension for input files | "../../../file.txt" |
| ${output.html} | The file extension for output files | "file.html" |
| ${pipeline, email address} | Email of user running the pipeline | "username@labkey.com" |
| ${pipeline, protocol description} | Provide a description | "" (an empty string is valid) |
| ${pipeline, protocol name} | The name of the protocol | "test" |
| ${pipeline, taskInfo} | See below | "../file-taskInfo.tsv" |
| ${pipeline, taskOutputParams} | Path to an output parameter file. See below | "file.params-out.tsv" |
| ${rLabkeySessionId} | Session ID for rLabKey | "LabKeyTransformSessionId" |
| ${sessionCookieName} | Session cookie name | "labkey.sessionCookieName = "LabKeyTransformSessionId"
labkey.sessionCookieContents = "3ba879ad8068932f4699a0310560f921495b71e91ae5133d722ac0a05c97c3ed"
" |
pipeline, taskInfo
Use the property ${pipeline, taskInfo} to provide the path to the taskInfo.tsv file containing current container, all input file paths, and all output file paths that LabKey writes out before executing every pipeline job. This property provides the file path; use it to load all the properties from the file.
pipeline, taskOutputParams
Use the property ${pipeline, taskOutputParams} to provide a path to an output parameter file. The output will be added to the recorded action's output parameters. Output parameters from a previous step are included as parameters (and token replacements) in subsequent steps.
Tasks
Tasks are defined in a LabKey Server module. They are file-based, so they can be created from scratch, cloned, exported, imported, renamed, and deleted. Tasks declare parameters, inputs, and outputs. Inputs may be files, parameters entered by users or by the API, a query, or a user selected set of rows from a query. Outputs may be files, values, or rows inserted into a table. Also, tasks may call other tasks.
File Operation Tasks
Exec Task
An example command line
.task.xml file that takes .hk files as input and writes .cms2 files:
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ExecTaskType" name="mytask" version="1.0">
<exec>
bullseye -s my.spectra -q ${q}
-o ${output.cms2} ${input.hk}
</exec>
</task>
Script Task
An example task configuration file that calls an R script:
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ScriptTaskType"
name="generateMatrix" version="0.0">
<description>Generate an expression matrix file (TSV format).</description>
<script file="RScript.r"/>
</task>
Inputs and Outputs
File inputs and outputs are identified by file extension. The input and output extensions cannot be the same. For example, the following configures the task to accept .txt files:
<inputs>
<file name="input.txt"/>
</inputs>
File outputs are automatically named using the formula: input file name + file extension set at <outputs><file name="output.tsv">. For example, If the input file is "myData1.txt", the output file will be named "myData1.tsv".
- The task name must be unique (no other task with the same name). For example: <task xmlns="http://labkey.org/pipeline/xml" name="myUniqueTaskName">
- An input must be declared, either implicitly or explicitly with XML configuration elements.
- Input and output files must not have the same file extensions. For example, the following is not allowed, because .tsv is declared for both input and output:
<inputs>
<file name="input.tsv"/>
</inputs>
<outputs>
<file name="output.tsv"/> <!-- WRONG - input and output cannot share the same file extension. -->
</outputs>
Configure required parameters with the attribute 'required', for example:
<inputs>
<file name="input.tsv"/>
<text name="param1" required="true"/>
</inputs>
Control the output location (where files are written) using the attributes outputDir or outputLocation.
Handling Multiple Input Files
Typically each input represents a single file, but sometimes a single input can represent multiple files. If your task supports more than one input file at a time, you can add the 'splitFiles' attribute to the input <file> declaration of the task. When the task is executed, the multiple input files will be listed in the
${pipeline, taskInfo} property file.
<inputs>
<file name="input.tsv" splitFiles="true"/>
</inputs>
Alternatively, if the entire pipeline should be executed once for every file, add the 'splittable' annotation to the top-level pipeline declaration. When the pipeline is executed, multiple jobs will be queued, one for each input file.
<pipeline xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
name="scriptset1-assayimport" version="0.0" splittable="true">
<tasks> … </tasks>
</pipeline>
Implicitly Declared Parameters, Inputs, and Outputs
Implicitly declared parameters, inputs, and outputs are allowed and identified by the dollar sign/curly braces syntax, for example, ${param1}.
- Inputs are identified by the pattern: ${input.XXX} where XXX is the desired file extension.
- Outputs are identified by the pattern: ${output.XXX} where XXX is the desired file extension.
- All others patterns are considered parameters: ${fooParam}, ${barParam}
For example, the following R script contains these implicit parameters:
- ${input.txt} - Input files have 'txt' extension.
- ${output.tsv} - Output files have 'tsv' extension.
- ${skip-lines} - An integer indicating how many initial lines to skip.
# reads the input file and prints the contents to stdout
lines = readLines(con="${input.txt}")
# skip-lines parameter. convert to integer if possible
skipLines = as.integer("${skip-lines}")
if (is.na(skipLines)) {
skipLines = 0
}
# lines in the file
lineCount = NROW(lines)
if (skipLines > lineCount) {
cat("start index larger than number of lines")
} else {
# start index
start = skipLines + 1
# print to stdout
cat("(stdout) contents of file: ${input.txt}\n")
for (i in start:lineCount) {
cat(sep="", lines[i], "n")
}
# print to ${output.tsv}
f = file(description="${output.tsv}", open="w")
cat(file=f, "# (output) contents of file: ${input.txt}\n")
for (i in start:lineCount) {
cat(file=f, sep="", lines[i], "n")
}
flush(con=f)
close(con=f)
}
Assay Database Import Tasks
The built-in task type
AssayImportRunTaskType looks for TSV and XLS files that were output by the previous task. If it finds output files, it uses that data to update the database, importing into whatever assay runs tables you configure.
An example task sequence file with two tasks: (1) generate a TSV file, (2) import that file to the database:
scriptset1-assayimport.pipeline.xml.
<pipeline xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
name="scriptset1-assayimport" version="0.0">
<!-- The description text is shown in the Import Data selection menu. -->
<description>Sequence: Call generateMatrix.r to generate a tsv file,
import this tsv file into the database. </description>
<tasks>
<!-- Task #1: Call the task generateMatrix (= the script generateMatrix.r) in myModule -->
<taskref ref="myModule:task:generateMatrix"/>
<!-- Task #2: Import the output/results of the script into the database -->
<task xsi:type="AssayImportRunTaskType" >
<!-- Target an assay by provider and protocol, -->
<!-- where providerName is the assay type -->
<!-- and protocolName is the assay design -->
<!-- <providerName>General</providerName> -->
<!-- <protocolName>MyAssayDesign</protocolName> -->
</task>
</tasks>
</pipeline>
The name attribute of the <pipeline> element, this must match the file name (minus the file extension). In this case: 'scriptset1-assayimport'.
The elements
providerName and
protocolName determine which runs table is targeted.
Pipeline Task Sequences / Jobs
Pipelines consist of a configured sequence of tasks. A "job" is a pipeline instance with specific input and outputs files and parameters. Task sequences are defined in files with the extension ".pipeline.xml".
Note the task references, for example "myModule:task:generateMatrix". This is of the form
<ModuleName>:task:<TaskName>, where <TaskName> refers to a task config file at /pipeline/tasks/<TaskName>.task.xml
An example pipeline file:
job1.pipeline.xml, which runs two tasks:
<pipeline xmlns="http://labkey.org/pipeline/xml"
name="job1" version="0.0">
<description> (1) Normalize and (2) generate an expression matrix file.</description>
<tasks>
<taskref ref="myModule:task:normalize"/>
<taskref ref="myModule:task:generateMatrix"/>
</tasks>
</pipeline>
Invoking Pipeline Sequences from the File Browser
Configured pipeline jobs/sequences can be invoked from the Pipeline File browser by selecting an input file(s) and clicking
Import Data. The list of available pipeline jobs is populated by the .pipeline.xml files. Note that the module(s) containing the script pipeline(s) must first be
enabled in the folder where you want to use them.
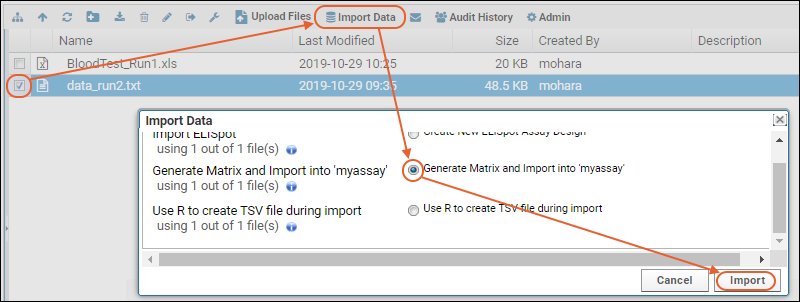
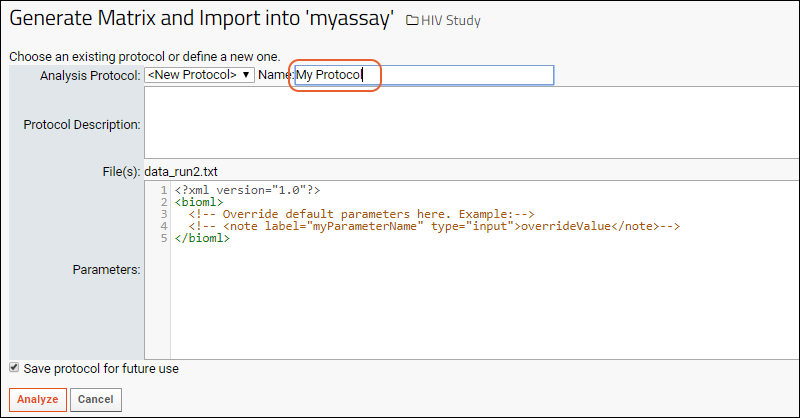
The server provides user interface for selecting/creating a protocol, for example, the job below is being run with the protocol "MyProtocol".
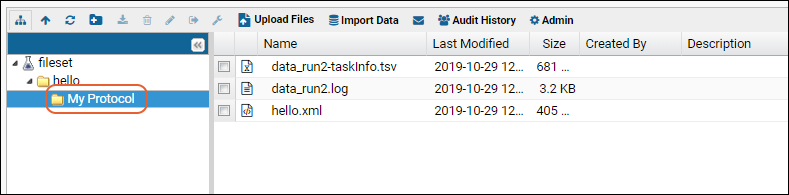
When the job is executed, the results will appear in a subfolder with the same name as the protocol selected.
Overriding Parameters
The default UI provides a panel for overriding default parameters for the job.
<?xml version="1.0" encoding="UTF-8"?>
<bioml>
<!-- Override default parameters here. -->
<note type="input" label="pipeline, protocol name">geneExpression1</note>
<note type="input" label="pipeline, email address">steveh@myemail.com</note>
</bioml>
Providing User Interface
You can override the default user interface by setting <analyzeURL> in the .pipeline.xml file.
<pipeline xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
name="geneExpMatrix-assayimport" version="0.0">
<description>Expression Matrix: Process with R, Import Results</description>
<!-- Overrides the default UI, user will see myPage.view instead. -->
<analyzeURL>/pipelineSample/myPage.view</analyzeURL>
<tasks>
...
</tasks>
</pipeline>
Invoking from JavaScript Clients
A
JavaScript wrapper (Pipeline.startAnalysis) is provided for invoking the script pipeline from JavaScript clients. For other script clients, such as Python and PERL, no wrapper is provided; but you can invoke script pipelines using an HTTP POST. See
below for details.
Example JavaScript that invokes a pipeline job through LABKEY.Pipeline.startAnalysis().
Note the value of taskId: 'myModule:pipeline:generateMatrix'. This is of the form
<ModuleName>:pipeline:<TaskName>, referencing a file at /pipeline/pipelines/<PipelineName>.pipeline.xml
function startAnalysis()
{
var protocolName = document.getElementById("protocolNameInput").value;
if (!protocolName) {
alert("Protocol name is required");
return;
}
var skipLines = document.getElementById("skipLinesInput").value;
if (skipLines < 0) {
alert("Skip lines >= 0 required");
return;
}
LABKEY.Pipeline.startAnalysis({
taskId: "myModule:pipeline:generateMatrix",
path: path,
files: files,
protocolName: protocolName,
protocolDescription: "",
jsonParameters: {
'skip-lines': skipLines
},
saveProtocol: false,
success: function() {
window.location = LABKEY.ActionURL.buildURL("pipeline-status", "showList.view")
}
});
}
Invoking from Python and PERL Clients
To invoke the script pipeline from Python and other scripting engines, use the HTTP API by sending an HTTP POST to pipeline-analysis-startAnalysis.api.
For example, the following POST invokes the Hello script pipeline shown
below, assuming that:
- there is a file named "test.txt" in the File Repository
- the Hello script pipeline is deployed in a module named "HelloWorld"
URL
pipeline-analysis-startAnalysis.api
- taskId takes the form <MODULE_NAME>:pipeline:<PIPELINE_NAME>, where MODULE_NAME refers to the module where the script pipeline is deployed.
- file is the file uploaded to the LabKey Server folder's file repository.
- path is relative to the LabKey Server folder.
- configureJson specifies parameters that can be used in script execution.
- protocolName is user input that determines the folder structure where log files and output results are written.
{
taskId: "HelloWorld:pipeline:hello",
file: ["test.txt"],
path: ".",
configureJson: '{"a": 3}',
protocolName: "helloProtocol",
saveProtocol: false
}LabKey provides a convenient
API testing page, for sending HTTP POSTs to invoke an API action. For example, the following screenshot shows how to use the API testing page to invoke the
HelloWorld script pipeline.

Execution Environment
When a pipeline job is run, a
job directory is created named after the job type and another child directory is created inside named after the protocol, for example, "create-matrix-job/protocol2". Log and output files are written to this child directory.
Also, while a job is running, a
work directory is created, for example, "run1.work". This includes:
- Parameter replaced script.
- Context 'task info' file with server URL, list of input files, etc.
If job is successfully completed, the work directory is cleaned up - any generated files are moved to their permanent locations, and the work directory is deleted.
Hello World R Script Pipeline
The following three files make a simple Hello World R script pipeline to include in a
HelloWorld module.
hello.r
# My first program in R Programming
myString <- "Hello, R World!"
print (myString)
# reads the input file and prints the contents
lines = readLines(con="${input.txt}")
print (lines)
hello.task.xml
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ScriptTaskType"
name="hello" version="0.0">
<description>Says hello in R.</description>
<inputs>
<file name="input.txt"/>
</inputs>
<script file="hello.r"/>
</task>
hello.pipeline.xml
<pipeline xmlns="http://labkey.org/pipeline/xml"
name="hello" version="0.0">
<description>Pipeline Job for Hello R.</description>
<tasks>
<taskref ref="HelloWorld:task:hello"/>
</tasks>
</pipeline>
Hello World Python Script Pipeline
The following three files make a simple Hello World Python script pipeline to include in a
HelloWorld module.
hello.py
print("A message from the Python script: 'Hello World!'")hellopy.task.xml
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ScriptTaskType"
name="hellopy" version="0.0">
<description>Says hello in Python.</description>
<inputs>
<file name="input.txt"/>
</inputs>
<script file="hello.py"/>
</task>
hellopy.pipeline.xml
<pipeline xmlns="http://labkey.org/pipeline/xml"
name="hellopy" version="0.0">
<description>Pipeline Job for Hello Python.</description>
<tasks>
<taskref ref="HelloWorld:task:hellopy"/>
</tasks>
</pipeline>
Hello World Python Script Pipeline - Inline Version
The following two files make a simple Hello World pipeline using an inline Python script to include in a
HelloWorld module.
hellopy-inline.task.xml
<task xmlns="http://labkey.org/pipeline/xml"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:type="ScriptTaskType"
name="hellopy-inline" version="0.0">
<description>Says hello in Python from an inline script.</description>
<inputs>
<file name="input.txt"/>
</inputs>
<script interpreter="py">print("A message from the inline Python script: 'Hello World!'")
</script>
</task>
hellopy-inline.pipeline.xml
<pipeline xmlns="http://labkey.org/pipeline/xml"
name="hellopy-inline" version="0.0">
<description>Pipeline Job for Hello Python - Inline version.</description>
<tasks>
<taskref ref="HelloWorld:task:hellopy-inline"/>
</tasks>
</pipeline>
Insert Assay Data Using the R API
You will find examples of using the R API to import assay results in the Rlabkey documentation. Syntax for including run level properties was added in version 3.4.3.
Other Resources
- LABKEY.Pipeline
- Pipeline XML Config Reference
- Example script pipelines from the LabKey Server automated tests. Download these .module files and deploy them to a production server, either through the server UI or by copying the .module files to <LABKEY_HOME>/externalModules and restarting the server. Once deployed, enable these modules in your folder.
Pipeline Task Listing
To assist in troubleshooting, you can find the full list of pipelines and tasks registered on your server via the Admin Console:
- Select > Site > Admin Console.
- Under Diagnostics, click Pipelines and Tasks.
Related Topics
 The server provides user interface for selecting/creating a protocol, for example, the job below is being run with the protocol "MyProtocol".
The server provides user interface for selecting/creating a protocol, for example, the job below is being run with the protocol "MyProtocol". When the job is executed, the results will appear in a subfolder with the same name as the protocol selected.
When the job is executed, the results will appear in a subfolder with the same name as the protocol selected.
